Summary
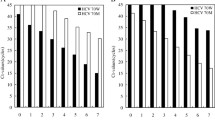
Since hepatitis C virus (HCV), a major causative agent of posttransfusional non-A, non-B hepatitis, is a positive stranded RNA virus, it is supposed to replicate via a negative RNA strand. Although strand specific reverse transcription-polymerase chain reaction (RT-PCR) method was recently developed to detect each strand of HCV RNA, the specificity of the strategy has remained to be determined. In this study, using in vitro transcribed positive and negative stranded HCV RNAs mixed with hepatic cellular RNA from normal liver, we found that this strategy did not distinguish between the two RNA strands, but that chemical modification of RNA samples at the 3′ end followed by strand specific RT-PCR made specific detection possible. Liver tissues, sera and peripheral blood mononuclear cells (PBMC) from ten patients with chronic HCV infection were analyzed with the novel strategy of RT-PCR combined with RNA modification. Positive and negative strands of HCV RNA were detected in liver tissues of ten (100%) and nine (90%) cases, respectively. Negative RNA strand was detected also in sera of five cases (50%), positive strand being detected in nine cases (90%). In PBMC, positive strand of HCV RNA was detected in eight cases (80%), whereas negative strand in only one case (10%), suggesting that HCV has much less cellular tropism to PBMC than to hepatocytes.
Similar content being viewed by others
References
Bouffard P, Hayashi PH, Acevedo R, Levy N, Zeldis JB (1992) Hepatitis C virus is detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells of infected patients. J Infect Dis 166: 1276–1280
Chomczynski P, Sacchi N (1987) Single step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159
Choo Q-L, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M (1989) Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244: 359–362
Cleaves GR, Ryan TE, Schlesinger RW (1981) Identification and characterization of type 2 dengue virus replicative intermediate and replicative form RNAs. Virology 111: 73–83
Fong T-L, Shindo M, Feinstone SW, Hoofnagle JH, Di Bisceglie AM (1991) Detection of replicative intermediates of hepatitis C viral RNA in liver and serum of patients with chronic hepatitis C. J Clin Invest 88: 1058–1060
Hu K-Q, Yu C-H, Vierling JM (1992) Direct detection of circulating hepatitis C virus RNA using probes from the 5′ untranslated region. J Clin Invest 89: 2040–2045
Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K (1990) Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA 87: 9524–9528
Kuo G, Choo Q-L, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE, Tegtmeier GE, Bonino F, Colombo M, Lee WS, Kuo C, Berger K, Shuster JR, Overby LR, Bradley DW, Houghton M (1989) An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244: 362–364
Lamas E, Baccarini P, Housset C, Kremsdorf D, Brechot C (1992) Detection of hepatitis C virus (HCV) RNA sequences in liver tissue by in situ hybridization. J Hepatol 16: 219–223
Maruyama HB, Hatanaka M, Gilden RV (1971) The 3′-terminal nucleotides of the high molecular weight RNA of C-type viruses. Proc Natl Acad Sci USA 68: 1999–2001
Miller RH, Purcell RH (1990) Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA 87: 2057–2061
Negro F, Pacchioni D, Shimizu Y, Miller RH, Bussolati G, Purcell RH, Bonio F (1992) Detection of intrahepatic replication of hepatitis C virus RNA by in situ hybridization and comparison with histopathology. Proc Natl Acad Sci USA 89: 2247–2251
Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH (1985) Nucleotide sequence of yellow fever virus: Implications for flavivirus gene expression and evolution. Science 229: 726–733
Takehara T, Hayashi N, Mita E, Hagiwara H, Ueda K, Katayama K, Kasahara A, Fusamoto H, Kamada T (1992) Detection of the minus strand of hepatitis C virus RNA by reverse transcription and polymerase chain reaction: Implication for hepatitis C virus replication in infected tissue. Hepatology 15: 387–390
Zignego AL, Macchia D, Monti M, Thiers V, Mazzetti M, Foschi M, Maggi E, Romagnani S, Gentilini P, Brechot C (1992) Infection of peripheral mononuclear blood cells by hepatitis C virus. J Hepatol 15: 382–386
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Gunji, T., Kato, N., Hijikata, M. et al. Specific detection of positive and negative stranded hepatitis C viral RNA using chemical RNA modification. Archives of Virology 134, 293–302 (1994). https://doi.org/10.1007/BF01310568
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01310568