Abstract
Myogenesis has been a leading model for elucidating the molecular mechanisms that underlie tissue differentiation and development since the discovery of MyoD. During myogenesis, the fate of myogenic precursor cells is first determined by Pax3/Pax7. This is followed by regulation of the myogenic differentiation program by muscle regulatory factors (Myf5, MyoD, Myog, and Mrf4) to form muscle tissues. Recent studies have uncovered a detailed myogenic program that involves the RP58 (Zfp238)-dependent regulatory network, which is critical for repressing the expression of inhibitor of DNA binding (Id) proteins. These novel findings contribute to a comprehensive understanding of the muscle differentiation transcriptional program.
Similar content being viewed by others
Introduction
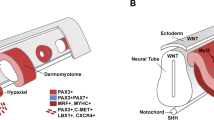
Vertebrate skeletal muscle is derived from the somites, which are established as paraxial mesoderm beside the neural tube and notochord (Fig. 1a) [1]. The somite progressively subdivides into two compartments, the dorsal dermomyotome and ventral sclerotome (Fig. 1b). The dorsal dermomyotome further splits into the dermatome and myotome in later stages of development to produce the trunk dermis and muscles, respectively. Myogenic precursor cells undergo an epithelial-mesenchymal transition (EMT) in response to signals from environmental cues, delaminate from the dermomyotome, and accumulate underneath to form the myotome (Fig. 1c) [2].
A schematic representation of somite patterning. a Each somite (S) forms beside the neural tube (NT) and notochord (N), adjacent to the lateral plate mesoderm (LPM). b The dorsal portion of the somite differentiates into the dermomyotome (DM), while the ventral quadrant gives rise to the sclerotome (SC). Signals from the floor plate and notochord (SHH) then specify the epaxial portion of the dermomyotome (yellow). Signals from the surface ectoderm (E; Wnt7a) and lateral plate mesoderm (BMP4) specify the hypaxial dermomyotome (green). c The two dermomyotome regions each produce a compartment of the myotome (M). The epaxial myotome forms from the epaxial lip of the dermomyotome (yellow), while the hypaxial myotome forms from the hypaxial lip (green)
The dermomyotome can also be divided into dorsal (epaxial) and ventral (hypaxial) portions, though no obvious morphological epaxial-hypaxial boundary exists. Sonic hedgehog (SHH) secreted from the floor plate and notochord specifies the epaxial dermomyotome, and gives rise to the epaxial myotome that forms the back muscles [3]. The hypaxial dermomyotome is specified by signals from the dorsal ectoderm (Wnt pathway) and the lateral plate mesoderm (Bmp4), and produces the hypaxial myotome that forms the limbs, diaphragm, and body wall muscles (Fig. 1b, c) [4].
All myogenic precursor cells in the dermomyotome express the paired-homeodomain transcription factor, Pax3 [2]. When myogenesis begins, Pax3 expression gradually decreases, and expression of basic helix-loop-helix (bHLH) transcription factors known as muscle regulatory factors (MRFs), such as Myf5, MyoD, Myog, and Mrf4, significantly increases [2]. These cells are then specified as myoblasts, fuse with each other, and finally differentiate into skeletal muscle fibers.
Gene targeting analysis using mouse models over the last 2 decades have shown that among these MRFs, Myf5 and MyoD are critical for myoblast determination and exhibit redundant functions. Although inactivation of either gene alone has no effect on skeletal muscle phenotype at the embryonic stage, Myf5/MyoD double knockout (DKO) mice have a complete lack of myoblasts and skeletal muscle throughout the body [5]. In contrast, Myog is a transcription factor that plays an essential role during muscle differentiation. Indeed, while myoblasts are specified normally in Myog KO mice, they cannot differentiate into myotubes [6, 7]. Myog is considered a direct downstream target of Myf5 and MyoD in the muscle gene network, and other MRFs cannot compensate for the defect [6, 7]. Mrf4 was first suggested to have a redundant function with Myog under the control of Myf5 and MyoD [8], but recent studies have shown that Mrf4 functions as a determinant of myogenic lineage in the early myotome (primary myogenesis) [9].
The early myotome possesses a distinct lineage that arises through the division of epaxial and hypaxial myotome regions. The epaxial myotome is formed by myoblasts derived from the dorsomedial lip of the dermomyotome. These myoblasts first express Myf5. On the other hand, the hypaxial myotome is formed by myoblasts derived from the ventrolateral lip of the dermomyotome. These myoblasts predominantly express MyoD (Fig. 1b). Though both Myf5 and MyoD single mutant mice show relatively normal muscle phenotypes, other abnormalities have been observed. Myf5−/− embryos show normal muscle development in the limb buds, but markedly delayed development of epaxial muscles. In contrast, while MyoD−/− mice display normal development of epaxial muscles, the development of limb muscles are significantly delayed. These results indicate that, even in the early myotome, at least two distinct myoblasts specified by the Myf5 and MyoD pathways exist and play different roles in muscle development [10].
Limb muscle development
While the hypaxial dermomyotome in the flank (inter-limb) elongates as epithelial sheets that give rise to body wall muscles and intercostal muscles, the hypaxial dermomyotome cells in the adjacent limb regions leave the epithelial structure after EMT and begin migrating towards their destination, i.e., the fore- and hind-limb bud mesenchyme [11]. These long-range migratory myogenic precursor cells express Pax3, retain their proliferative state, and do not express MRFs during the migration (Fig. 2a). After they reach the target destination, dorsal and ventral muscle-forming regions in the limb bud begin to express MRFs, downregulate Pax3, and finally differentiate into muscle fibers [2]. Splotch mutant mice, which lack functional Pax3, are devoid of all limb muscles because of defects in the migration of precursors from the somites to the limb bud [12].
a Pax3 expression in the mouse embryo. Pax3 mRNA is not detected in the hindlimb at E9.5. The dashed line outlines the hindlimb bud. Arrows mark Pax3 (+) cells that have migrated into the limbs of the E10.5 embryo. The right panel is an enlargement of the central panel. Labeled structures are the dorsomedial lip of the dermomyotome (DML), ventrolateral lip of the dermomyotome (VLL), forelimb (FL), hindlimb (HL), neural tube (NT), heart (H), and nasal pit (N). b Schematic representation of the expression patterns of genes involved in dermomyotome development. Blue cells indicate myogenic precursor cells migrating to the limb bud. c A model of genes responsible for the development and migration of myogenic precursor cells to the limb bud. The migrating myogenic precursors delaminate from the VLL and populate in the dorsal (dmm) and ventral muscle masses (vmm) in the limb bud. Six1/4, Eya1/2, and Pax3 are indispensable for the specification of myogenic precursor cells in the ventrolateral lip of the dermomyotome. c-Met is important for the delamination of VLL cells. Migrating myogenic precursor cells express Pax3, c-Met, Lbx1, CXCR4, Sp5, and Pitx2. Muscle differentiation within the limb is controlled by the expression of the MRFs, Pitx3, and RP58
The key molecules involved in EMT of these myogenic precursor cells are hepatocyte growth factor/scatter factor (HGF/SF) and its receptor, c-Met [11]. These migratory myogenic precursor cells in the hypaxial lip of the dermomyotome express c-Met in addition to Pax3 [11, 12] (Fig. 2B). In contrast, HGF/SF mRNA is predominantly expressed in the adjacent limb mesenchyme [11, 13]. Both c-Met−/− and HGF/SF−/− mice have muscle-free limbs, resembling the Splotch phenotype, due to the lack of myogenic precursor cell migration from the dermomyotome to the limb [11, 14]. c-Met is considered a downstream target of Pax3 because the Splotch mutant shows a loss of c-Met expression in the hypaxial dermomyotome [12]. However, it is unclear whether c-Met is a direct Pax3 target in vivo [15] and if these c-Met (+) precursor cells have a chemotactic response to HGF/SF [11, 13, 16].
Long-range migratory myogenic precursor cells are generated only in the occipital, cervical, and fore- and hindlimb levels of the dermomyotome ventrolateral lip. These precursors specifically express the Lbx1 homeobox transcription factor (Fig. 2b) [17]. Lbx1 inactivation leads to the lack of dorsal muscle mass in the forelimb and all muscles in the hindlimb. In this mutant, c-Met (+) migrating precursors are correctly specified and delaminate from the ventrolateral lip of the dermomyotome, but cannot enter laterally into the limb bud [11]. These results suggest that Lbx1 is important for migratory myogenic precursor cells to find the correct guidance cues to the limb bud. It is not known why precursors of the ventrolateral lip of the inter-limb region, which express both Pax3 and c-Met, can exclude Lbx1 expression.
Other genes expressed in migratory myogenic cells
Six family transcription factors (Six1, Six4) and Eya transcriptional cofactors (Eya1, Eya2) are also critical for hypaxial muscle specification and the migration of hypaxial myogenic precursor cells from the somites to the limb. In both Six1/Six4 and Eya1/Eya2 DKO mice, Pax3 is absent from the hypaxial dermomyotome, resulting in severe defects of precursor cell migration into the limb bud (Fig. 2c) [18, 19]. This indicates that Six and Eya proteins function as upstream regulators of Pax3 during the establishment of the hypaxial dermomyotome.
Sp5 is expressed in the dorsomedial and ventrolateral lips of the dermomyotome, overlapping with the expression pattern of Pax3 [20]. Sp5 mRNA is also detected in migrating myogenic precursor cells from the dermomyotome to the limb, and its expression is not detected in Splotch mice, suggesting that Sp5 is expressed in migratory muscle precursors and its expression is regulated by Pax3 [20] (Fig. 2c).
Pitx2 and Pitx3 are also expressed in limb muscles (Fig. 2c) [21]. Pitx2 is expressed in migratory myogenic precursor cells, whereas Pitx3 can be seen during muscle differentiation. Pitx3 mutant mice show upregulated and prolonged Pitx2 expression, suggesting that Pitx2 and Pitx3 may be partially redundant [21].
Recent studies have shown that the chemokine receptor CXCR4 is also expressed in migratory myogenic precursor cells (Fig. 2c) [22]. The CXCR4 ligand, stromal derived factor 1 (Sdf1), is specifically expressed in the middle mesenchyme of the limb bud, which is the final destination of these migratory cells. Ectopic Sdf1 expression in the chick limb bud causes an aberrant accumulation of CXCR4 (+) myogenic cells near the source. In CXCR4−/− mice, fewer progenitor cells reach the limb bud and apoptosis increases, suggesting an important role of CXCR4 for migratory cues and cell survival [22]. CXCR4 genetically interacts with Gab1, the adaptor molecule that mediates interaction with the c-Met receptor. In CXCR4/Gab1 DKO mice, the number of migratory myogenic cells reaching the forelimb is significantly reduced compared to each single mutant, suggesting that a threshold number of precursor cells reaching the limb is necessary to compensate for muscle defects [22].
PAX transcription factors and the specification of myogenic precursor cells in gastrulation and adults
Myogenic precursor cells in the dermomyotome also express Pax7, the analog of Pax3 [2]. Pax3 positive precursor cells accumulate in the dorsomedial and ventrolateral lips of the dermomyotome during development (Fig. 2a, b), while Pax7 expression is more prominent in the central region of the dermomyotome [23, 24].
In the adult body, muscle growth and regeneration events depend on the proliferation and differentiation of muscle stem cells, termed satellite cells, which are located under the basal lamina of muscle fibers. These satellite cells express Pax7 and are mitotically quiescent in undamaged muscle. Upon injury, they are activated, migrate to the injured area, proliferate, and fuse with each other to replace degenerated muscle fibers. This is accompanied by upregulation of MRFs and the downregulation of Pax7. In Pax7−/− mice, satellite cells progressively die by apoptosis after birth, and Pax3 cannot compensate [25]. Recent findings suggest that Pax3 is also expressed in many, but not all, satellite cells [25, 26]. Quiescent satellite cells also express the receptor c-Met, and the ligand HGF/SF is a trigger for satellite cell activation [27]. These combined results indicate that Pax transcription factors are important for the specification and maintenance of muscle progenitor cells both in early somites and in adult muscle tissues, and suggest that a very similar transcriptional network is orchestrated during embryonic skeletal muscle formation and adult muscle regeneration.
A population of skeletal muscle progenitor cells resides in the central dermomyotome that expresses both Pax3 and Pax7 [28, 29]. These Pax3(+)Pax7(+) cells are mitotically active, do not co-express MRFs after entering the myotome, and persist in all fetal and neonatal muscles [23, 24, 26]. Pax3/Pax7 DKO mice show some early embryonic muscles of the myotome, but completely lack further muscle development because of loss of the muscle progenitor cell population [26]. Cells failing to express Pax3 or Pax7 either die or assume a non-myogenic fate. These results indicate that Pax3(+)Pax7(+) mitotic progenitor cells are responsible for fetal muscle growth (secondary myogenesis) and contribute to satellite cells of postnatal mice, suggesting that embryonic muscle progenitors and satellite cells are commonly derived from the central dermomyotome [24, 26].
MyoD and muscle differentiation
Weintraub’s discovery of MyoD in 1986 shed new light on the molecular nature of skeletal muscle differentiation [30]. Weintraub’s group cloned MyoD by subtractive hybridization from azacytidine-treated myoblasts (by subtracting cDNA of azacytidine-induced myoblasts from mouse 10T1/2 fibroblasts) and demonstrated that MyoD alone was enough to convert 10T1/2 fibroblasts into myoblasts [31].
MyoD (as wells as other bHLH MRF factors) binds the E-box sequence (CANNTG) in promoters of downstream muscle target genes, thereby driving the transcription of these muscle-related genes in collaboration with myocyte enhancer factor 2 (Mef2) transcription factors (Fig. 3a) [32]. Mef2C is specifically expressed in muscle tissues and binds to DNA near E-boxes. Normally, these bHLH MRFs dimerize with E-proteins (E12, E47 and HEB), which are ubiquitous throughout the body, to activate downstream gene expression [32]. However, in the presence of inhibitor of DNA binding (Id) proteins (Id1–Id4), which are negative regulators of myogenesis, MyoD cannot activate the transcription of downstream target genes [33]. Ids can heterodimerize with E-proteins (and to a lesser extent with MyoD), thereby attenuating MyoD function (Fig. 3a). Although the molecular mechanisms regulating the inhibition of Id protein expression are unknown, we recently found that the transcriptional repressor RP58 (mouse Zfp238) is expressed in early differentiated muscle tissue and represses Id2/Id3 expression, allowing MyoD to promote muscle differentiation (Fig. 3b) [34]. RP58 is first expressed in limb muscle tissues around E11.5, coincident with Myog, and like Myog, is upregulated by acetylated MyoD during early differentiation stages [34, 35]. These results indicate that MyoD both activates (via Myog) and represses (via RP58) a distinct gene set that permits the progression of skeletal myogenesis to late differentiation (Fig. 3c) [34, 36].
MyoD activates target gene expression. a MyoD dimerizes with E2A and binds to the E-box in promoters of muscle-specific genes. Id proteins prevent the formation of MyoD/E2A heterodimers and their DNA-binding activities. b RP58 represses Id2 and Id3 transcription by binding to the putative RP58 binding site in the Id2 and Id3 promoters. c Proposed myogenesis regulatory network via RP58
Future directions
Expression of the aristaless-like homeobox transcription factor Alx4 demarcates the central region of the dermomyotome, excluding the dorsomedial and ventrolateral lips [37]. En1 expression partially overlaps with Alx4 expression in the epaxial portion of the central dermomyotome and is adjacent to expression of the bHLH transcription factor Sim1, which is expressed in the hypaxial portion of the central dermomyotome that overlaps with Alx4 [37]. Using fate mapping analysis in the mouse embryo, Atit et al. [38] demonstrated that En1-expressing central dermomyotomal cells have three distinct fates: dorsal dermis, epaxial muscle, and, unexpectedly, some interscapular brown fat. In addition, Ben-Yair et al. [39] showed that a single cell in the central dermomyotome has the potential to produce both myotome and dermatome cells by asymmetric cell division. It is noted that En1 and Sim1 are expressed not only in the dermomyotome, but also in the adjacent myotome and dermis (Fig. 4). These results imply that multipotent common progenitor cells that reside in the central dermomyotome region can also express Sim1. Moreover, these central dermomyotome cells are also Pax3 and Pax7 positive in early development (Fig. 2b, 4). It was previously shown that Pax3 and Pax7 positive cells derived from the central dermomyotome are also found in adult muscle satellite cells beside the myotome, indicating that muscle satellite cells have a common origin with embryonic myogenic precursor cells in the dermomyotome of somites [24, 26]. Therefore, En1 may contribute to the specification of adult muscle satellite cells, although its expression in these cells has not been reported.
En1 and Sim1 are broadly expressed in the epaxial and hypaxial compartments in the somites, respectively. Pax7 (+) and Pax3 (+) cells predominantly reside in the central portion of the dermomyotome, overlapping with Alx4 expression, and retain this expression even after migrating to the myotome (M). DML dorsomedial lip, VLL ventrolateral lip of the dermomyotome
Recent studies have shown that brown fat cells are derived from skeletal muscle lineage progenitor cells. It was previously thought that white fat cells and brown fat cells have a common adipogenic origin. However, through in vivo cell tracing analysis, Seale et al. showed that brown fat cells are derived from Myf5 (a skeletal muscle-specific transcription factor) positive muscle cells during early development [40]. They also showed that the SET domain-containing Zinc finger transcription factor Prdm16 determines the fate of these multipotent Myf5 positive precursors to become brown fat cells. This ability of Prdm16 is bidirectional. Prdm16 knockdown in brown fat cells results in skeletal muscle-specific gene upregulation, and its overexpression in C2 muscle cells causes downregulation of muscle genes and brown fat formation. Furthermore, Tseng et al. [41] found that Bmp7 can stimulate the brown adipose lineage, activating Prdm16, Pgc-1a, and the brown fat marker, uncoupling protein 1 (Ucp1). Bmp7−/− mice show a dramatic reduction in brown fat and an almost complete lack of Ucp1. In contrast, Myog−/− mutant mice show ectopic brown adipocytes in some regions that are normally skeletal muscle tissue [6]. This myoblast/brown adipocyte precursor may exhibit some plasticity for cell fate and can switch fates based on surrounding cues. This is very intriguing given the ability of brown adipose tissue to burn fat. However, it is currently unknown whether there are interactions between Prdm16 and Bmp7 expressing regions within the somites and central dermomyotome where brown adipocytes are produced. Further studies of the genetic interactions between Prdm16/Bmp7 and genes expressed in the central dermomyotome, such as Alx4, En1, Sim1, Pax transcription factors, and other unknown factors, will shed light on mechanisms underlying the specification of different cell fates of central dermomyotome cells.
References
Christ B, Ordahl CP (1995) Early stages of chick somite development. Anat Embryol (Berl) 191(5):381–396
Yusuf F, Brand-Saberi B (2006) The eventful somite: patterning, fate determination and cell division in the somite. Anat Embryol (Berl) 211(Suppl 1):21–30. doi:10.1007/s00429-006-0119-8
Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, Emerson CP Jr (1999) Sonic hedgehog controls epaxial muscle determination through myf5 activation. Development 126(18):4053–4063
Cheng L, Alvares LE, Ahmed MU, El-Hanfy AS, Dietrich S (2004) The epaxial-hypaxial subdivision of the avian somite. Dev Biol 274(2):348–369. doi:10.1016/j.ydbio.2004.07.020
Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R (1993) Myod or myf-5 is required for the formation of skeletal muscle. Cell 75(7):1351–1359. doi:0092-8674(93)90621-V
Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH (1993) Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364(6437):501–506. doi:10.1038/364501a0
Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I (1993) Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364(6437):532–535. doi:10.1038/364532a0
Zhang W, Behringer RR, Olson EN (1995) Inactivation of the myogenic bhlh gene mrf4 results in up-regulation of myogenin and rib anomalies. Genes Dev 9(11):1388–1399
Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S (2004) Mrf4 determines skeletal muscle identity in myf5:Myod double-mutant mice. Nature 431(7007):466–471. doi:10.1038/nature02876
Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA (1997) Myod and myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development 124(23):4729–4738
Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C (1999) The role of sf/hgf and c-met in the development of skeletal muscle. Development 126(8):1621–1629
Daston G, Lamar E, Olivier M, Goulding M (1996) Pax-3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development 122(3):1017–1027
Heymann S, Koudrova M, Arnold H, Koster M, Braun T (1996) Regulation and function of sf/hgf during migration of limb muscle precursor cells in chicken. Dev Biol 180(2):566–578. doi:S0012160696903292
Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C (1995) Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376(6543):768–771. doi:10.1038/376768a0
Epstein JA, Shapiro DN, Cheng J, Lam PY, Maas RL (1996) Pax3 modulates expression of the c-met receptor during limb muscle development. Proc Natl Acad Sci USA 93(9):4213–4218
Takayama H, La Rochelle WJ, Anver M, Bockman DE, Merlino G (1996) Scatter factor/hepatocyte growth factor as a regulator of skeletal muscle and neural crest development. Proc Natl Acad Sci USA 93(12):5866–5871
Schafer K, Braun T (1999) Early specification of limb muscle precursor cells by the homeobox gene lbx1h. Nat Genet 23(2):213–216. doi:10.1038/13843
Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P (2005) Six1 and six4 homeoproteins are required for pax3 and mrf expression during myogenesis in the mouse embryo. Development 132(9):2235–2249. doi:10.1242/dev.01773
Grifone R, Demignon J, Giordani J, Niro C, Souil E, Bertin F, Laclef C, Xu PX, Maire P (2007) Eya1 and eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol 302(2):602–616. doi:10.1016/j.ydbio.2006.08.059
Harrison SM, Houzelstein D, Dunwoodie SL, Beddington RS (2000) Sp5, a new member of the sp1 family, is dynamically expressed during development and genetically interacts with brachyury. Dev Biol 227(2):358–372. doi:10.1006/dbio.2000.9878
L’Honore A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, Gage P, Camper S, Drouin J (2007) Sequential expression and redundancy of pitx2 and pitx3 genes during muscle development. Dev Biol 307(2):421–433. doi:10.1016/j.ydbio.2007.04.034
Vasyutina E, Stebler J, Brand-Saberi B, Schulz S, Raz E, Birchmeier C (2005) Cxcr4 and gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev 19(18):2187–2198. doi:10.1101/gad.346205
Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S (2005) Pax3/pax7 mark a novel population of primitive myogenic cells during development. Genes Dev 19(12):1426–1431. doi:10.1101/gad.345505
Gros J, Manceau M, Thome V, Marcelle C (2005) A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435(7044):954–958. doi:10.1038/nature03572
Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M (2006) Pax3 and pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol 172(1):91–102. doi:10.1083/jcb.200508044
Relaix F, Rocancourt D, Mansouri A, Buckingham M (2005) A pax3/pax7-dependent population of skeletal muscle progenitor cells. Nature 435(7044):948–953. doi:10.1038/nature03594
Allen RE, Sheehan SM, Taylor RG, Kendall TL, Rice GM (1995) Hepatocyte growth factor activates quiescent skeletal muscle satellite cells in vitro. J Cell Physiol 165(2):307–312. doi:10.1002/jcp.1041650211
Ben-Yair R, Kalcheim C (2005) Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development 132(4):689–701. doi:10.1242/dev.01617
Lagha M, Brunelli S, Messina G, Cumano A, Kume T, Relaix F, Buckingham ME (2009) Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev Cell 17(6):892–899. doi:10.1016/j.devcel.2009.10.021
Lassar AB, Paterson BM, Weintraub H (1986) Transfection of a DNA locus that mediates the conversion of 10t1/2 fibroblasts to myoblasts. Cell 47(5):649–656. doi:0092-8674(86)90507-6
Davis RL, Weintraub H, Lassar AB (1987) Expression of a single transfected cdna converts fibroblasts to myoblasts. Cell 51(6):987–1000. doi:0092-8674(87)90585-X
Puri PL, Sartorelli V (2000) Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol 185(2):155–173. doi:10.1002/1097-4652(200011)185:2<155:AID-JCP1>3.0.CO;2-Z
Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61(1):49–59. doi:0092-8674(90)90214-Y
Yokoyama S, Ito Y, Ueno-Kudoh H, Shimizu H, Uchibe K, Albini S, Mitsuoka K, Miyaki S, Kiso M, Nagai A, Hikata T, Osada T, Fukuda N, Yamashita S, Harada D, Mezzano V, Kasai M, Puri PL, Hayashizaki Y, Okado H, Hashimoto M, Asahara H (2009) A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor rp58. Dev Cell 17(6):836–848. doi:10.1016/j.devcel.2009.10.011
Di Padova M, Caretti G, Zhao P, Hoffman EP, Sartorelli V (2007) Myod acetylation influences temporal patterns of skeletal muscle gene expression. J Biol Chem 282(52):37650–37659. doi:10.1074/jbc.M707309200
Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD (2005) An initial blueprint for myogenic differentiation. Genes Dev 19(5):553–569. doi:10.1101/gad.1281105
Ahmed MU, Cheng L, Dietrich S (2006) Establishment of the epaxial-hypaxial boundary in the avian myotome. Dev Dyn 235(7):1884–1894. doi:10.1002/dvdy.20832
Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA (2006) Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol 296(1):164–176. doi:10.1016/j.ydbio.2006.04.449
Ben-Yair R, Kahane N, Kalcheim C (2003) Coherent development of dermomyotome and dermis from the entire mediolateral extent of the dorsal somite. Development 130(18):4325–4336
Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM (2008) Prdm16 controls a brown fat/skeletal muscle switch. Nature 454(7207):961–967. doi:10.1038/nature07182
Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR (2008) New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454(7207):1000–1004. doi:10.1038/nature07221
Acknowledgments
We thank all members of the Asahara laboratory for their critical discussions and reading of the manuscript. This work was supported by a Grant-in-Aid for Scientific Research (MEXT), Health and Labour Science Research Grants.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Yokoyama, S., Asahara, H. The myogenic transcriptional network. Cell. Mol. Life Sci. 68, 1843–1849 (2011). https://doi.org/10.1007/s00018-011-0629-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-011-0629-2