Abstract
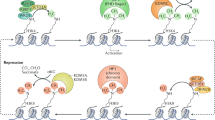
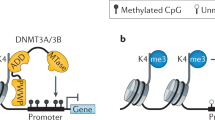
Histone modifications contribute to the precise regulation of transcription by recruiting non-histone proteins and controlling chromatin conformation. These covalent modifications are dynamically regulated by many enzymes that modify histones at specific residues in different ways. Histone modifiers contribute to development as well as cellular responses to extracellular stimuli. Mutations in the genes encoding them cause various diseases, including developmental disorders and certain malignancies. Haploinsufficiency for some histone methyltransferases, one of the principal modifiers of the histone modification network, are associated with particular congenital diseases, including Sotos syndrome, Wolf–Hirschhorn syndrome, and 9q syndrome. In this review, we discuss the molecular function of the histone methyltransferases and the human diseases associated with their dysfunction.

Similar content being viewed by others
References
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Baker LA, Allis CD, Wang GG (2008) PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res, Fundam Mol Mech Mutagen 647:3–12
Klose RJ, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8:307–318
Sims RJ, Reinberg D (2006) Histone H3 Lys 4 methylation: caught in a bind? Genes Dev 20:2779–2786
Maurer-Stroh S, Dickens NJ, Hughes-Davies L et al (2003) The tudor domain ‘royal family’: tudor, plant agenet, chromo, PWWP and MBT domains. Trends Biochem Sci 28:69–74
Bhaumik SR, Smith E, Shilatifard A (2007) Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 14:1008–1016
Barski A, Cuddapah S, Cui K et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Mikkelsen TS, Ku M, Jaffe DB et al (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560
Heintzman ND, Stuart RK, Hon G et al (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318
Wang P, Lin C, Smith ER et al (2009) Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29:6074–6085
Milne TA, Briggs SD, Brock HW et al (2002) MLL targets set domain methyltransferase activity to hox gene promoters. Mol Cell 10:1107–1117
Steward MM, Lee J, O’Donovan A et al (2006) Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 13:852–854
Wysocka J, Myers MP, Laherty CD et al (2003) Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 17:896–911
Gregory GD, Vakoc CR, Rozovskaia T et al (2007) Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol 27:8466–8479
Hamamoto R, Furukawa Y, Morita M et al (2004) SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol 6:731–740
Chuikov S, Kurash JK, Wilson JR et al (2004) Regulation of p53 activity through lysine methylation. Nature 432:353–360
Tan X, Rotllant J, Li H et al (2006) SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci USA 103:2713–2718
Gottlieb PD, Pierce SA, Sims RJ et al (2002) Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet 31:25–32
Vermeulen M, Mulder KW, Denissov S et al (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 Lysine 4. Cell 131:58–69
Shi X, Hong T, Walter KL et al (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442:96–99
Peña PV, Davrazou F, Shi X et al (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442:100–103
Kim K, Geng L, Huang S (2003) Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res 63:7619–7623
Matsui T, Leung D, Miyashita H et al (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927–931
Kim J, Daniel J, Espejo A et al (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7:397–430
Karagianni P, Amazit L, Qin J et al (2008) ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol 28:705–717
Iwase S, Lan F, Bayliss P et al (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128:1077–1088
Jensen L, Amende M, Gurok U et al (2005) Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet 76:227–236
Cao R, Wang L, Wang H et al (2002) Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298:1039–1043
Fiskus W, Wang Y, Sreekumar A et al (2009) Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood 114:2733–2743
Simon JA, Lange CA (2008) Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res, Fundam Mol Mech Mutagen 647:21–29
Tan J, Yang X, Zhuang L et al (2007) Pharmacologic disruption of polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 21:1050–1063
Miranda TB, Cortez CC, Yoo CB et al (2009) DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther 8:1579–1588
Rayasam GV, Wendling O, Angrand PO et al (2003) NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J 22:3153–3163
Nimura K, Ura K, Shiratori H et al (2009) A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome. Nature 460:287–291
Brown MA, Sims RJ, Gottlieb PD et al (2006) Identification and characterization of Smyd 2: a split SET/MYND domain-containing histone H 3 lysine 36-specific methyltransferase that interacts with the Sin 3 histone deacetylase complex. Mol Cancer 5:26
Edmunds JW, Mahadevan LC, Clayton AL (2007) Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27:406–420
Sun X, Wei J, Wu X et al (2005) Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280:35261–35271
Luco RF, Pan Q, Tominaga K et al (2010) Regulation of alternative splicing by histone modifications. Science 327:996–1000
Zhang P, Du J, Sun B et al (2006) Structure of human MRG 15 chromo domain and its binding to Lys 36-methylated histone H 3. Nucleic Acids Res 34:6621–6628
Carrozza MJ, Li B, Florens L et al (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592
Keogh MC, Kurdistani SK, Morris SA et al (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593–605
Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6:838–849
Botuyan MV, Lee J, Ward IM et al (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127:1361–1373
Huyen Y, Zgheib O, DiTullio RA Jr et al (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406–411
Okada Y, Feng Q, Lin Y et al (2005) hDOT1L links histone methylation to leukemogenesis. Cell 121:167–178
Jones B, Su H, Bhat A et al (2008) The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4:e1000190
Nishioka K, Rice JC, Sarma K et al (2002) PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9:1201–1213
Schotta G, Lachner M, Sarma K et al (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18:1251–1262
Marango J, Shimoyama M, Nishio H et al (2008) The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood 111:3145–3154
Oda H, Okamoto I, Murphy N et al (2009) Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29:2278–2295
Trojer P, Li G, Sims RJ III et al (2007) L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129:915–928
Krause CD, Yang Z, Kim Y et al (2007) Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther 113:50–87
Bedford MT, Richard S (2005) Arginine methylation: an emerging regulator of protein function. Mol Cell 18:263–272
Kirmizis A, Santos-Rosa H, Penkett CJ et al (2009) Distinct transcriptional outputs associated with mono- and dimethylated histone H3 arginine 2. Nat Struct Mol Biol 16:449–451
Guccione E, Bassi C, Casadio F et al (2007) Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449:933–937
Hyllus D, Stein C, Schnabel K et al (2007) PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev 21:3369–3380
Iberg AN, Espejo A, Cheng D et al (2008) Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem 283:3006–3010
Li B, Carey M, Workman J (2007) The role of chromatin during transcription. Cell 128:707–719
Huang N, vom Baur E, Garnier J et al (1998) Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J 17:3398–3412
Angrand P, Apiou F, Stewart AF et al (2001) NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 74:79–88
Kurotaki N, Imaizumi K, Harada N et al (2002) Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30:365–366
Tatton-Brown K, Rahman N (2006) Sotos syndrome. Eur J Hum Genet 15:264–271
Cerveira N, Correia C, Dória S et al (2003) Frequency of NUP98–NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia 17:2244–2247
Rosati R, La Starza R, Veronese A et al (2002) NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t (8;11)(p11.2;p15). Blood 99:3857–3860
Wang GG, Cai L, Pasillas MP et al (2007) NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 9:804–812
Berdasco M, Ropero S, Setien F et al (2009) Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci 106:21830–21835
Li Y, Trojer P, Xu C et al (2009) The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem 284:34283–34295
Nielsen AL, Jorgensen P, Lerouge T et al (2004) Nizp1, a novel multitype zinc finger protein that interacts with the NSD1 histone lysine methyltransferase through a unique C2HR motif. Mol Cell Biol 24:5184–5196
Lu T, Jackson MW, Wang B et al (2010) Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci 107:46–51
Maas NMC, Van Buggenhout G, Hannes F et al (2008) Genotype-phenotype correlation in 21 patients with Wolf–Hirschhorn syndrome using high resolution array comparative genome hybridisation (CGH). J Med Genet 45:71–80
Bergemann A, Cole F, Hirschhorn K (2005) The etiology of Wolf–Hirschhorn syndrome. Trends Genet 21:188–195
Zollino M, Lecce R, Fischetto R et al (2003) Mapping the Wolf–Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am J Hum Genet 72:590–597
Wright T, Ricke D, Denison K et al (1997) A transcript map of the newly defined 165 kb Wolf–Hirschhorn syndrome critical region. Hum Mol Genet 6:317–324
Dimmer KS, Navoni F, Casarin A et al (2008) LETM1, deleted in Wolf–Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum Mol Genet 17:201–214
Simon R, Bergemann AD (2008) Mouse models of Wolf–Hirschhorn syndrome. Am J Med Genet C Semin Med Genet 148C:275–280
Chesi M, Nardini E, Lim RS et al (1998) The t (4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92:3025–3034
Stec I, Wright TJ, van Ommen GJ et al (1998) WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf–Hirschhorn syndrome critical region and is fused to IgH in t (4; 14) multiple myeloma. Hum Mol Genet 7:1071–1082, Published erratum appears in Hum Mol Genet 1998 Sep; 7 (9): 1527
Kang HB, Choi Y, Lee JM et al (2009) The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett 583:1880–1886
Kleefstra T, Brunner HG, Amiel J et al (2006) Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet 79:370–377
Kleefstra T, Smidt M, Banning M et al (2005) Disruption of the gene euchromatin histone methyl transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet 42:299–306
Balemans MC, Huibers MM, Eikelenboom NW et al (2010) Reduced exploration, increased anxiety, and altered social behavior: autistic-like features of euchromatin histone methyltransferase 1 heterozygous knockout mice. Behav Brain Res 208:47–55
Tachibana M, Ueda J, Fukuda M et al (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19:815–826
Schaefer A, Sampath SC, Intrator A et al (2009) Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64:678–691
Collins RE, Northrop JP, Horton JR et al (2008) The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol 15:245–250
Fritsch L, Robin P, Mathieu JR et al (2010) A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37:46–56
Ueda J, Tachibana M, Ikura T et al (2006) Zinc finger protein wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem 281:20120–20128
Acknowledgment
We thank John A McGrath, Lo Wan Ning, Christine Vogler, and Masaki Mori for their critical reading of the manuscript. This work was supported by grants from Japan Heart Foundation Research Grant and Kanae foundation for the promotion of medical science to K. N.
Disclosure of potential conflict of interests
The authors declare no conflict of interests related to this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nimura, K., Ura, K. & Kaneda, Y. Histone methyltransferases: regulation of transcription and contribution to human disease. J Mol Med 88, 1213–1220 (2010). https://doi.org/10.1007/s00109-010-0668-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-010-0668-4