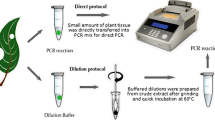
Abstract
Vegetative insecticidal protein (Vip), a unique class of insecticidal protein, is now part of transgenic plants for conferring resistance against lepidopteron pests. In order to address the imminent regulatory need for detection and labeling of vip3A carrying genetically modified (GM) products, we have developed a standard single PCR and a multiplex PCR assay. As far as we are aware, this is the first report on PCR-based detection of a vip3A-type gene (vip-s) in transgenic cotton and tobacco. Our assay involves amplification of a 284-bp region of the vip-s gene. This assay can possibly detect as many as 20 natural wild-type isolates bearing a vip3A-like gene and two synthetic genes of vip3A in transgenic plants. The limit of detection as established by our assay for GM trait (vip-s) is 0.1%. Spiking with nontarget DNA originating from diverse plant sources had no inhibitory effect on vip-s detection. Since autoclaving of vip-s bearing GM leaf samples showed no deterioration/interference in detection efficacy, the assay seems to be suitable for processed food products as well. The vip-s amplicon identity was reconfirmed by restriction endonuclease assay. The primer set for vip-s was equally effective in a multiplex PCR assay format (duplex, triplex and quadruplex), used in conjunction with the primer sets for the npt-II selectable marker gene, Cauliflower mosaic virus 35S promoter and nopaline synthetase terminator, enabling concurrent detection of the transgene, regulatory sequences and marker gene. Further, the entire transgene construct was amplified using the forward primer of the promoter and the reverse primer of the terminator. The resultant amplicon served as a template for nested PCR to confirm the construct integrity. The method is suitable for screening any vip3A-carrying GM plant and food. The availability of a reliable PCR assay method prior to commercial release of vip3A-based transgenic crops and food would facilitate rapid and efficient regulatory compliance.









Similar content being viewed by others
References
Estruch JJ, Warren GW, Mullins MA, Nye GJ, Craig JA, Koziel MG (1996) Proc Natl Acad Sci USA 93:5389–5394
GMO Crop Database (2007) AGBIOS, Merrickville. http://www.agbios.com/dbase.php
Rang C, Gil P, Neisner N, Rie JV, Frutos R (2005) Appl Environ Microbiol 71:6276–6281
Crickmore N (2005) Vip proteins. http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/viptree.pdf
Lee MK, Miles P, Chen JS (2006) Biochem Biophys Res Commun 339:1043–1047
International Centre for Genetic Engineering and Biotechnology (2007) Raj Kamal Bhatnagar, Group Leader Plant Biology: Insect Resistance. http://www.icgeb.org/RESEARCH/ND/Bhatnagar.htm
James C (2005) ISAAA briefs. Brief 34. Global status of commercialized biotech/GM crops: 2005. ISAAA, Ithaca. http://www.isaaa.org/Resources/publications/briefs/34/download/isaaa-brief-34-2005.pdf
Ahmed FE (2002) Trends Biotechnol 20:215–223
Environmental Protection Agency (2005) Bacillus thuringiensis VIP3A protein and the genetic material necessary for its production; temporary exemption from the requirement of a tolerance. http://www.epa.gov/fedrgstr/EPA-PEST/2005/April/Day-28/p8530.htm
Porcar M, Juarez-Perez V (2003) FEMS Microbiol Rev 26:419–432
Rice WC (1999) Appl Microbiol Lett 28:378–382
Germini A, Zanetti A, Salati C, Rossi S, Forré C, Schmid S, Marchelli R (2004) J Agric Food Chem 52:3275–3280
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 1.32–1.34
Lipp M, Bluth A, Eyquem F, Kruse L, Schimmel H, Van den Eede G, Anklam E (2001) Eur Food Res Technol 212:497–504
European Committee for Standardization (2001) European standard document no CEN/TC 275/WG 11 N149
GMO Methods Database (2007) European Commission, DG Joint Research Centre, Institute for Health and Consumer Protection, Ispra. http://bgmo.jrc.ec.europa.eu/home/ict/methodsdatabase.htm
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Plant Mol Biol 17:1105–1109
Arumuganathan K, Earle ED (1991) Plant Mol Biol Rep 9:208–218
Laura B, Petra H, Simon K, Van den Eede G (2001) Review of GMO detection and quantification techniques. European Commission, Joint Research Centre, Institute for Health and Consumer Protection, Food Products and Consumer Goods Unit, Ispra. http://www.osservaogm.it/pdf/JRCReview.pdf
Kay S, Van den Eede G (2001) Nat Biotechnol 19:405
Warren GW, Koziel MG, Mullins MA, Nye GJ, Carr B, Desai WM, Kostochka K, Duck NB, Estruch JJ (1996) Patent WO 96/10083. World Intellectual Property Organization
Office of the Gene Technology Regulator (2007) Record of GMOs and GM products—licences involving an intentional release of GMOs into the environment. http://www.ogtr.gov.au/gmorec/ir.htm
Sharma M, Charak KS, Ramanaiah TV (2003) Curr Sci 84:297–302
Holst-Jensen A, Ronning SB, Lovseth A, Berdal KG (2003) Anal Bioanal Chem 375:985–993
Alexander TW, Sharma R, Deng MY, Whetsell AJ, Jennings JC, Wang Y, Okine E, Damgaard D, McAllister TA (2004) J Biotechnol 112:255–266
Yin Z, Plader W, Malepszy S (2004) J Appl Genet 45:127–144
Herrera S (2005) Nat Biotechnol 23:514
Acknowledgements
This work was supported by a research grant from the Department of Biotechnology, India. The authors thank Sameer Sawant, National Botanical Research Institute, Lucknow, for meaningful discussions and critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
An application for an Indian patent (1891/DEL2006/17.08.07) comprising a substantive part of this study has been filed.
ITRC communication no. 2516.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, C.K., Ojha, A., Bhatanagar, R.K. et al. Detection and characterization of recombinant DNA expressing vip3A-type insecticidal gene in GMOs—standard single, multiplex and construct-specific PCR assays. Anal Bioanal Chem 390, 377–387 (2008). https://doi.org/10.1007/s00216-007-1714-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-007-1714-0