Abstract
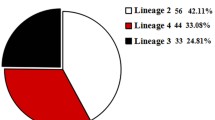
In this study, we aimed to identify the genetic lineages of Mycobacterium tuberculosis isolates in Isfahan via the mycobacterial interspersed repetitive-unit-variable number tandem repeat typing method based on 15 loci. Forty-nine M. tuberculosis isolates were collected between 2013 and 2015 from Tuberculosis patients in Mollahadi Sabzevari Tuberculosis Center in Isfahan. All isolates were typed by 15-locus MIRU-VNTR typing. The highest percentage of isolates, 44.89 % (22/49), belonged to the Euro-American lineage, while the frequencies of the East-African–Indian, East-Asian, and Indo-Oceanic lineages were 28.57 % (14/49), 24.4 % (12/49), and 2.04 % (1/49), respectively. Among the 22 isolates of the Euro-American lineage, those belonging to the NEW-1 sub-lineage were most prevalent (24.4 %). Approximately, the same proportion of isolates belonging to the Delhi/CAS, Beijing, and NEW-1 sub-lineages were identified in Iranian and Afghan immigrant patients. The Delhi/CAS and Beijing sub-lineage isolates were prevalent among patients who had been previously treated for TB. Results showed that all of the 49 MIRU-VNTR patterns were unique and the clustering rate of the 15-locus MIRU-VNTR was 0.0 (minimum recent transmission). The results of this study show that the lineages of M. tuberculosis isolates in Isfahan are similar to those reported in the Eastern Mediterranean region (indicative of the epidemiological relationship between the countries in the region). The low clustering rate in our results reveals that transmission of tuberculosis in Isfahan is, in most cases, a reactivation of previous tuberculosis infection and the role of recently transmitted disease is minor.



Similar content being viewed by others
References
Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S (2008) Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 46(8):2692–2699
Bouklata N, Supply P, Jaouhari S, Charof R, Seghrouchni F, Sadki K, El Achhab Y, Nejjari C, Filali-Maltouf A, Lahlou O (2015) Molecular Typing of Mycobacterium tuberculosis complex by 24-locus based MIRU-VNTR typing in conjunction with spoligotyping to assess genetic diversity of strains circulating in Morocco. PLoS ONE 10(8):e0135695
Buu TN, Huyen MN, Lan NN, Quy HT, Hen NV, Zignol M, Borgdorff MW, Dv Soolingen, Cobelens FG (2009) Mycobacterium tuberculosis genotype and case notification rates, rural Vietnam, 2003–2006. Emerg Infect Dis 15(10):1570–1577
Chamla D, Nie S, Duan Q (2004) Retrospective descriptive study of adult tuberculosis in Wuhan, China. Int J Tuberc Lung Dis 8(6):730–736
Click ES, Moonan PK, Winston CA, Cowan LS, Oeltmann JE (2012) Relationship between Mycobacterium tuberculosis phylogenetic lineage and clinical site of tuberculosis. Clin Infect Dis 54(2):211–219
Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G (2013) Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 45(10):1176–1182
Cowan LS, Diem L, Monson T, Wand P, Temporado D, Oemig TV, Crawford JT (2005) Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol 43(2):688–695
Eisenach KD, Cave MD, Bates JH, Crawford JT (1990) Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis 161(5):977–981
Elisseeff V (2000) The silk roads. Highways of culture and commerce. Berghahn, New York
Filliol I, Driscoll JR, van Soolingen D, Kreiswirth BN, Kremer K, Valétudie G, Anh DD, Barlow R, Banerjee D, Bifani PJ (2003) Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J Clin Microbiol 41(5):1963–1970
Fitzgibbon M, Gibbons N, Roycroft E, Jackson S, O’Donnell J, O’Flanagan D, Rogers T (2013) A snapshot of genetic lineages of Mycobacterium tuberculosis in Ireland over a two-year period, 2010 and 2011. Euro Surveill 18(3):1–7
Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D (2002) Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis 8(8):843–849
González Díaz A, Battaglioli T, Díaz Rodríguez R, Goza Valdés R, González Ochoa E, van der Stuyft P (2015) Molecular epidemiology of tuberculosis in Havana, Cuba, 2009. Trop Med Int Health 20(11):1534–1542
Hasan Z, Tanveer M, Kanji A, Hasan Q, Ghebremichael S, Hasan R (2006) Spoligotyping of Mycobacterium tuberculosis isolates from Pakistan reveals predominance of Central Asian Strain 1 and Beijing isolates. J Clin Microbiol 44(5):1763–1768
Hashemi A, Shojaei H, Heidarieh P, Aslani MM, Naser AD (2012) Genetic diversity of Iranian clinical isolates of Mycobacterium tuberculosis. New Microbiol 35(1):61–65
Itah AY, Udofia SM (2005) Epidemiology and endemicity of pulmonary tuberculosis (PTB) in Southeastern Nigeria. Southeast Asian J Trop Med Public Health 36(2):317
Moradi M, Arababadi MK, Hassanshahi G (2008) Tuberculosis in the Afghan immigrant in Kerman province of Iran. J Biol Sci 8(6):1107–1109
Mustafa Ali R, Trovato A, Couvin D, Al-Thwani AN, Borroni E, Dhaer FH, Rastogi N, Cirillo DM (2014) Molecular epidemiology and genotyping of Mycobacterium tuberculosis isolated in Baghdad. Biomed Res Int. doi:10.1155/2014/580981
Osman DA, Phelippeau M, Drancourt M, Musso D (2015) Diversity of Mycobacterium tuberculosis lineages in French Polynesia. J Microbiol Immunol Infect. doi:10.1016/j.jmii.2015.05.018
Palittapongarnpim P, Luangsook P, Tansuphaswadikul S, Chuchottaworn C, Prachaktam R, Sathapatayavongs B (1997) Restriction fragment length polymorphism study of Mycobacterium tuberculosis in Thailand using IS6110 as probe. Int J Tuberc Lung Dis 1(4):370–376
Pang Y, Zhou Y, Zhao B, Liu G, Jiang G, Xia H, Song Y, Shang Y, Wang S, Y-l Zhao (2012) Spoligotyping and drug resistance analysis of Mycobacterium tuberculosis strains from national survey in China. PLoS ONE 7(3):e32976
Ramazanzadeh R, Sayhemiri K (2014) Prevalence of Beijing family in Mycobacterium tuberculosis in world population: systematic review and meta-analysis. Int J Mycobact 3(1):41–45
Ramazanzadeh R, Farnia P, Amirmozafari N, Ghazi F, Ghadertotonchi Z, Kamran J, Mohammadi F, Mirsaedi M, Masjedi M (2006) Comparison between molecular epidemiology, geographical regions and drug resistance in Mycobacterium tuberculosis strains isolated from Iranian and Afghan patients. Chemotherapy 52(6):316–320
Ramazanzadeh R, Farnia P, Amirmozafari N (2009) Characterization of Mycobacterium tuberculosis complex isolated from Iranian and Afghani patients by spoligotyping method. Braz J Microbiol 40(2):314–320
Roetzer A, Schuback S, Diel R, Gasau F, Ubben T, di Nauta A, Richter E, Rüsch-Gerdes S, Niemann S (2011) Evaluation of Mycobacterium tuberculosis typing methods in a 4-year study in Schleswig-Holstein, Northern Germany. J Clin Microbiol 49(12):4173–4178
Schaaf H, Shean K, Donald P (2003) Culture confirmed multidrug resistant tuberculosis: diagnostic delay, clinical features, and outcome. Arch Dis Child 88(12):1106–1111
Serkani JE, Isfahani BN, Safaei HG, Kermanashahi R, Asghari G (2012) Evaluation of the effect of Humulus lupulus alcoholic extract on rifampin-sensitive and resistant isolates of Mycobacterium tuberculosis. Res pharm Sci 7(4):235–242
Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK (1994) The epidemiology of tuberculosis in San Francisco—a population-based study using conventional and molecular methods. N Engl J Med 330(24):1703–1709
Soini H, Pan X, Amin A, Graviss EA, Siddiqui A, Musser JM (2000) Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J Clin Microbiol 38(2):669–676
Sola C, Filliol I, Legrand E, Lesjean S, Locht C, Supply P, Rastogi N (2003) Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol 3(2):125–133
Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S (2006) Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44(12):4498–4510
Torkaman MRA, Nasiri MJ, FaRnia P, Shahhosseiny MH, Mozafari M, Velayati AA (2014) Estimation of recent transmission of Mycobacterium tuberculosis strains among Iranian and Afghan immigrants: a cluster-based study. J Clin Diagn Res 8(9):05. doi:10.7860/JCDR/2014/8886.4864
Toungoussova OS, Sandven P, Mariandyshev AO, Nizovtseva NI, Bjune G, Caugant DA (2002) Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J Clin Microbiol 40(6):1930–1937
Van Soolingen D, Qian L, De Haas P, Douglas JT, Traore H, Portaels F, Qing HZ, Enkhsaikan D, Nymadawa P, Van Embden J (1995) Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol 33(12):3234–3238
Warren R, Victor T, Streicher E, Richardson M, Van Der Spuy G, Johnson R, Chihota V, Locht C, Supply P, Van Helden P (2004) Clonal expansion of a globally disseminated lineage of Mycobacterium tuberculosis with low IS6110 copy numbers. J Clin Microbiol 42(12):5774–5782
WHO (2015) Global tuberculosis report 2015. WHO, Geneva
Zhang J, Heng S, Le Moullec S, Refregier G, Gicquel B, Sola C, Guillard B (2011) A first assessment of the genetic diversity of Mycobacterium tuberculosis complex in Cambodia. BMC Infect Dis 11(1):42
Acknowledgments
This study was funded by the Isfahan University of Medical Sciences with grant number 191123.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Riyahi Zaniani, F., Moghim, S., Mirhendi, H. et al. Genetic Lineages of Mycobacterium tuberculosis Isolates in Isfahan, Iran. Curr Microbiol 74, 14–21 (2017). https://doi.org/10.1007/s00284-016-1145-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1145-2