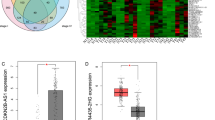
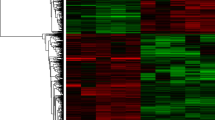
Abstract
Recently, accumulating evidence has demonstrated that non-coding RNAs (ncRNAs) play a vital role in oncogenicity. Nevertheless, the regulatory mechanisms and functions remain poorly understood, especially for lncRNAs and circRNAs. In this study, we simultaneously detected, for the first time, the expression profiles of the whole transcriptome, including miRNA, circRNA and lncRNA + mRNA, in five pairs of laryngeal squamous cell carcinoma (LSCC) and matched non-carcinoma tissues by microarrays. Five miRNAs, four circRNAs, three lncRNAs and five mRNAs that were dysregulated were selected to confirm the verification of the microarray data by quantitative real-time PCR (qRT-PCR) in 20 pairs of LSCC samples. We constructed LSCC-related competing endogenous RNA (ceRNA) networks of lncRNAs and circRNAs (circRNA or lncRNA–miRNA–mRNA) respectively. Functional annotation revealed the lncRNA-mediated ceRNA network were enriched for genes involved in the tumor-associated pathways. Hsa_circ_0033988 with the highest degree in the circRNA-mediated ceRNA network was associated with fatty acid degradation, which was responsible for the depletion of fat in tumor-associated cachexia. Finally, to clarify the ncRNA co-regulation mechanism, we constructed a circRNA–lncRNA co-regulated network by integrating the above two networks and identified 9 modules for further study. A subnetwork of module 2 with the most dysregulated microRNAs was extracted to establish the ncRNA-involved TGF-β-associated pathway. In conclusion, our findings provide a high-throughput microarray data of the coding and non-coding RNAs and establish the foundation for further functional research on the ceRNA regulatory mechanism of non-coding RNAs in LSCC.






Similar content being viewed by others
References
Anastasiadou E, Jacob LS, Slack FJ (2018) Non-coding RNA networks in cancer. Nat Rev Cancer 18:5–18. https://doi.org/10.1038/nrc.2017.99
Bierie B, Moses HL (2006) Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer 6:506–520. https://doi.org/10.1038/nrc1926
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Burkhart DL, Sage J (2008) Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8:671–682. https://doi.org/10.1038/nrc2399
Caldas C, Brenton JD (2005) Sizing up miRNAs as cancer genes. Nat Med 11:712–714. https://doi.org/10.1038/nm0705-712
Chew CL, Conos SA, Unal B, Tergaonkar V (2018) Noncoding RNAs: master regulators of inflammatory signaling. Trends Mol Med 24:66–84. https://doi.org/10.1016/j.molmed.2017.11.003
Cosetti M, Yu GP, Schantz SP (2008) Five-year survival rates and time trends of laryngeal cancer in the US population. Arch Otolaryngol Head Neck Surg 134:370–379. https://doi.org/10.1001/archotol.134.4.370
Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr (2013) Cellular fatty acid metabolism and cancer. Cell Metab 18:153–161. https://doi.org/10.1016/j.cmet.2013.05.017
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y, Sun B, Wang Y, Wang L, Huang Q (2017) Circular RNAs expression profiles in human gastric cancer. Sci Rep 7:9060. https://doi.org/10.1038/s41598-017-09076-6
de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20:1453–1454. https://doi.org/10.1093/bioinformatics/bth078
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3
Derynck R, Zhang YE (2003) Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425:577–584. https://doi.org/10.1038/nature02006
Dittrich A, Gautrey H, Browell D, Tyson-Capper A (2014) The HER2 signaling network in breast cancer—like a spider in its web. J Mammary Gland Biol Neoplasia 19:253–270. https://doi.org/10.1007/s10911-014-9329-5
Dong Y, Sui L, Sugimoto K, Tai Y, Tokuda M (2001) Cyclin D1-CDK4 complex, a possible critical factor for cell proliferation and prognosis in laryngeal squamous cell carcinomas. Int J Cancer 95:209–215
Du L, Li H, Zhu C, Zheng R, Zhang S, Chen W (2015) Incidence and mortality of laryngeal cancer in China, 2011. Chin J Cancer Res 27:52–58. https://doi.org/10.3978/j.issn.1000-9604.2015.02.02
Edefonti V, Bravi F, Garavello W, la Vecchia C, Parpinel M, Franceschi S, Dal Maso L, Bosetti C, Boffetta P, Ferraroni M, Decarli A (2010) Nutrient-based dietary patterns and laryngeal cancer: evidence from an exploratory factor analysis. Cancer Epidemiol Biomark Prev 19:18–27. https://doi.org/10.1158/1055-9965.EPI-09-0900
Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105. https://doi.org/10.1101/gr.082701.108
Fu BM, Liu Y (2012) Microvascular transport and tumor cell adhesion in the microcirculation. Ann Biomed Eng 40:2442–2455. https://doi.org/10.1007/s10439-012-0561-0
Genden EM, Ferlito A, Silver CE, Jacobson AS, Werner JA, Suárez C, Leemans CR, Bradley PJ, Rinaldo A (2007) Evolution of the management of laryngeal cancer. Oral Oncol 43:431–439. https://doi.org/10.1016/j.oraloncology.2006.08.007
Gene Ontology C et al (2013) Gene ontology annotations and resources. Nucleic Acids Res 41:D530–D535. https://doi.org/10.1093/nar/gks1050
Graf F, Mosch B, Koehler L, Bergmann R, Wuest F, Pietzsch J (2010) Cyclin-dependent kinase 4/6 (cdk4/6) inhibitors: perspectives in cancer therapy and imaging. Mini Rev Med Chem 10:527–539
Guo W, Giancotti FG (2004) Integrin signalling during tumour progression. Nat Rev Mol Cell Biol 5:816–826. https://doi.org/10.1038/nrm1490
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:409. https://doi.org/10.1186/s13059-014-0409-z
Hu B, Cai H, Zheng R, Yang S, Zhou Z, Tu J (2017) Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int J Biochem Cell Biol 92:34–42. https://doi.org/10.1016/j.biocel.2017.09.008
Hwang HW, Mendell JT (2007) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 96(Suppl):R40–R44
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31:e15–e115
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Lai Y (2017) A statistical method for the conservative adjustment of false discovery rate (q-value). BMC Bioinformatics 18:69. https://doi.org/10.1186/s12859-017-1474-6
Lee YS, Dutta A (2006) MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs 7:560–564
Li JQ, Yang J, Zhou P, Le YP, Gong ZH (2015) The biological functions and regulations of competing endogenous RNA. Yi Chuan 37:756–764. https://doi.org/10.16288/j.yczz.15-073
Li LJ, Leng RX, Fan YG, Pan HF, Ye DQ (2017) Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs. Exp Cell Res 361:1–8. https://doi.org/10.1016/j.yexcr.2017.10.010
Lopez-Romero P, Gonzalez MA, Callejas S, Dopazo A, Irizarry RA (2010) Processing of Agilent microRNA array data. BMC Res Notes 3:18. https://doi.org/10.1186/1756-0500-3-18
Malumbres M, Barbacid M (2009) Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9:153–166. https://doi.org/10.1038/nrc2602
Mattick JS (2011) Long noncoding RNAs in cell and developmental biology. Semin Cell Dev Biol 22:327. https://doi.org/10.1016/j.semcdb.2011.05.002
McAllister SS, Weinberg RA (2014) The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 16:717–727. https://doi.org/10.1038/ncb3015
McLaughlin F, Finn P, La Thangue NB (2003) The cell cycle, chromatin and cancer: mechanism-based therapeutics come of age. Drug Discov Today 8:793–802
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S (2017) Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform 18:780–788. https://doi.org/10.1093/bib/bbw053
Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, Bader GD, Ferrin TE (2011) clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12:436. https://doi.org/10.1186/1471-2105-12-436
Ni T, He Z, Dai Y, Yao J, Guo Q, Wei L (2017) Oroxylin A suppresses the development and growth of colorectal cancer through reprogram of HIF1alpha-modulated fatty acid metabolism. Cell Death Dis 8:e2865. https://doi.org/10.1038/cddis.2017.261
Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W (2015) ceRNA in cancer: possible functions and clinical implications. J Med Genet 52:710–718. https://doi.org/10.1136/jmedgenet-2015-103334
Qu J, Li M, Zhong W, Hu C (2015) Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med 8:17110–17116
Ryden M et al (2008) Lipolysis—not inflammation, cell death, or lipogenesis—is involved in adipose tissue loss in cancer cachexia. Cancer 113:1695–1704. https://doi.org/10.1002/cncr.23802
Sahu A, Singhal U, Chinnaiyan AM (2015) Long noncoding RNAs in cancer: from function to translation. Trends Cancer 1:93–109. https://doi.org/10.1016/j.trecan.2015.08.010
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358. https://doi.org/10.1016/j.cell.2011.07.014
Sanchez-Mejias A, Tay Y (2015) Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol 8:30. https://doi.org/10.1186/s13045-015-0129-1
Shi Q, Chen YG (2017) Interplay between TGF-beta signaling and receptor tyrosine kinases in tumor development. Sci China Life Sci 60:1133–1141. https://doi.org/10.1007/s11427-017-9173-5
Shuwen H, Qing Z, Yan Z, Xi Y (2018) Competitive endogenous RNA in colorectal cancer: a systematic review. Gene 645:157–162. https://doi.org/10.1016/j.gene.2017.12.036
Slaby O, Laga R, Sedlacek O (2017) Therapeutic targeting of non-coding RNAs in cancer. Biochem J 474:4219–4251. https://doi.org/10.1042/BCJ20170079
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX, Wu ZH, Chen XW, Gao P, Miao ZF, Wang ZN (2017) Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun 8:289. https://doi.org/10.1038/s41467-017-00304-1
Tay Y, Rinn J, Pandolfi PP (2014) The multilayered complexity of ceRNA crosstalk and competition. Nature 505:344–352. https://doi.org/10.1038/nature12986
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121. https://doi.org/10.1073/pnas.091062498
Wangyang Z, Daolin J, Yi X, Zhenglong L, Lining H, Yunfu C, Xingming J (2018) NcRNAs and cholangiocarcinoma. J Cancer 9:100–107. https://doi.org/10.7150/jca.21785
Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R (2018) Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci 75:467–484. https://doi.org/10.1007/s00018-017-2626-6
Yang Z, Xie L, Han L, Qu X, Yang Y, Zhang Y, He Z, Wang Y, Li J (2017) Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics 7:3106–3117. https://doi.org/10.7150/thno.19016
Zhang YE (2017) Non-Smad signaling pathways of the TGF-beta family. Cold Spring Harb Perspect Biol 9: a022129. https://doi.org/10.1101/cshperspect.a022129
Zhang SJ, Chen X, Li CP, Li XM, Liu C, Liu BH, Shan K, Jiang Q, Zhao C, Yan B (2017a) Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci 58:6500–6509. https://doi.org/10.1167/iovs.17-22698
Zhang Y, Liang W, Zhang P, Chen J, Qian H, Zhang X, Xu W (2017b) Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res 36:152. https://doi.org/10.1186/s13046-017-0624-z
Zheng S, Zheng D, Dong C, Jiang J, Xie J, Sun Y, Chen H (2017) Development of a novel prognostic signature of long non-coding RNAs in lung adenocarcinoma. J Cancer Res Clin Oncol 143:1649–1657. https://doi.org/10.1007/s00432-017-2411-9
Zhong Y, du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W (2018) Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 17:79. https://doi.org/10.1186/s12943-018-0827-8
Funding
This study was supported by the National Natural Science Foundation of China (No. 81572647).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was approved by the Human Research Ethics Committee from Harbin Medical University. Informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human tissues were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Electronic supplementary material
Online Resource 1:
The microarray original data of miRNA, circRNA and lncRNA+mRNA in five pairs of LSCC and matched non-carcinoma tissues (XLSX 31608 kb)
Online Resource 2:
Primers used for qRT-PCR. (PDF 82 kb)
Online Resource 3:
The top 30 significant terms by GO and KEGG pathway analysis for differential expressed mRNAs in the LSCC microarray. (PDF 489 kb)
Online Resource 4:
Volcano plots for the expression profiles of miRNAs(A), lncRNAs(B), mRNAs(C) and circRNAs(D-F). (PDF 827 kb)
Online Resource 5:
The expression correlation between microarray and PCR. (PDF 967 kb)
Online Resource 6:
The top 10 ranked lncRNAs according to degree in the lncRNA-miRNA-mRNA ceRNA network. (PDF 158 kb)
Online Resource 7:
The sub-ceRNA-network of LINC00657. (PDF 256 kb)
Online Resource 8:
KEGG pathway analysis of the differentially expressed mRNAs in the LINC00657 subnetwork (PDF 182 kb)
Online Resource 9:
GO and KEGG pathway enrichment analysis of the differentially expressed mRNAs in the circRNA-miRNA-mRNA ceRNA network. (PDF 406 kb)
Online Resource 10:
The top 10 circRNAs sorted by degree in the circRNA-miRNA-mRNA ceRNA network (PDF 85 kb)
Online Resource 11:
KEGG pathway enrichment analysis of the differentially expressed mRNAs in the circRNA-lncRNA co-regulated ceRNA network. (PDF 191 kb)
Online Resource 12:
The detailed information of nine modules in the circRNA-lncRNA co-regulated ceRNA network. (PDF 432 kb)
Online Resource 13:
KEGG pathway enrichment analysis of the differentially expressed mRNAs in module 2. (PDF 909 kb)
Rights and permissions
About this article
Cite this article
Zhao, R., Li, FQ., Tian, LL. et al. Comprehensive analysis of the whole coding and non-coding RNA transcriptome expression profiles and construction of the circRNA–lncRNA co-regulated ceRNA network in laryngeal squamous cell carcinoma. Funct Integr Genomics 19, 109–121 (2019). https://doi.org/10.1007/s10142-018-0631-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-018-0631-y