Abstract
Introduction
The development of common forms of diabetes comes from the interaction between environmental and genetic factors, and the consequences of poor glycemic control in these patients could result in several complications. Metabolomic studies for type 2 diabetes mellitus in serum/plasma have reported changes in numerous metabolites, which might be considered possible targets for future mechanistic research. However, the specific role of a particular metabolite as cause or consequence of diabetes derangements is difficult to establish.
Objectives
As type 2 diabetes is a disease that changes the metabolic profile in several levels, this work aimed to compare the metabolomic profiles of type 2 diabetes mellitus and non-diabetic participants. In addition, we exploited our family-based study design to bring a better understanding of the causal relationship of identified metabolites and diabetes.
Methods
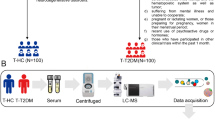
In the current study, population based metabolomics was applied in 939 subjects from the Baependi Heart Study. Participants were separated into two groups: diabetic (77 individuals) and non-diabetic (862 individuals), and the metabolic profile was performed by GC/MS technique.
Results
We have identified differentially concentrated metabolites in serum of diabetic and non-diabetic individuals. We identified 72 metabolites up-regulated in type 2 diabetes mellitus compared to non-diabetics. It was possible to recapitulate the main pathways that the literature shows as changed in diabetes. Also, based on metabolomic profile, we separated pre-diabetic individuals (with glucose concentration between 100–125 mg/dL) from non-diabetics and diabetics. Finally, using heritability analysis, we were able to suggest metabolites in which altered levels may precede diabetic development.
Conclusion
Our data can be used to derive a better understanding of the causal relationship of the observed associations and help to prioritize diabetes-associated metabolites for further work.




Similar content being viewed by others
References
Chevalier, S., Marliss, E. B., Morais, J. A., Lamarche, M., & Gougeon, R. (2005). Whole-body protein anabolic response is resistant to the action of insulin in obese women. The American Journal of Clinical Nutrition, 82(2), 355–365.
de Oliveira, C. M., Pereira, A. C., de Andrade, M., Soler, J. M., & Krieger, J. E. (2008). Heritability of cardiovascular risk factors in a Brazilian population: Baependi heart study. BMC Medical Genetics, 9, 32. doi:10.1186/1471-2350-9-32.
Dieterle, F., Ross, A., Schlotterbeck, G., & Senn, H. (2006). Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. application in 1H NMR metabonomics. Analytical Chemistry, 78(13), 4281–4290. doi:10.1021/ac051632c.
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 6(7), 1060–1083. doi:10.1038/nprot.2011.335.
Fernstrom, J. D. (2005). Branched-chain amino acids and brain function. The Journal of Nutrition, 135, 1539S–1546S.
Fiehn, O. (2002). Metabolomics—The link between genotypes and phenotypes. Plant Molecular Biology, 48, 155–171.
Fiehn, O., Garvey, W. T., Newman, J. W., Lok, K. H., Hoppel, C. L., & Adams, S. H. (2010). Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS One, 5(12), e15234. doi:10.1371/journal.pone.0015234.
Gall, W. E., Beebe, K., Lawton, K. A., Adam, K.-P., Mitchell, M. W., Nakhle, P. J., et al. (2010). Alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One, 5(5), e10883. doi:10.1371/journal.pone.0010883.
Horimoto, A. R. V. R., Giolo, S. R., Oliveira, C. M., Alvim, R. O., Soler, J. P., de Andrade, M., et al. (2011). Heritability of physical activity traits in Brazilian families: The Baependi Heart Study. BMC Medical Genetics, 12(1), 155. doi:10.1186/1471-2350-12-155.
Mikaelyan, N. P., Nguen, H. Z., & Terent’ev, A. A. (2015). Metabolic abnormalities in erythrocyte membranes and liver tissue homogenates in experimental diabetes mellitus. Bulletin of Experimental Biology and Medicine, 159(2), 197–200. doi:10.1007/s10517-015-2921-x.
Newgard, C. B., An, J., Bain, J. R., Muehlbauer, M. J., Stevens, R. D., Lien, L. F., et al. (2013). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism, 9(4), 311–326. doi:10.1016/j.cmet.2009.02.002.A.
Padberg, I., Peter, E., González-Maldonado, S., Witt, H., Mueller, M., Weis, T., et al. (2014). A new metabolomic signature in type-2 diabetes mellitus and its pathophysiology. PLoS One, 9(1), e85082. doi:10.1371/journal.pone.0085082.
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature, 490(7418), 55–60. doi:10.1038/nature11450.
Shah, S. H., Crosslin, D. R., Haynes, C. S., Nelson, S., Turer, C. B., Stevens, R. D., et al. (2012). Branched-chain amino acid levels are associated with improvement in insulin resistance with weight loss. Diabetologia, 55(2), 321–330. doi:10.1007/s00125-011-2356-5.
Sharma, A., Chavali, S., Mahajan, A., Tabassum, R., Banerjee, V., Tandon, N., et al. (2005). Genetic association, post-translational modification, and protein-protein interactions in type 2 diabetes mellitus. Molecular & Cellular Proteomics: MCP, 4(8), 1029–1037. doi:10.1074/mcp.M500024-MCP200.
Suhre, K. (2014). Metabolic profiling in diabetes. The Journal of Endocrinology, 221(3), R75–R85. doi:10.1530/JOE-14-0024.
Swan, J. W., Anker, S. D., Walton, C., Godsland, I. F., Clark, A. L., Leyva, F., et al. (1997). Insulin resistance in chronic heart failure: Relation to severity and etiology of heart failure. Journal of the American College of Cardiology, 30(2), 527–532. doi:10.1016/S0735-1097(97)00185-X.
Trifonova, O., Lokhov, P., & Archakov, A. (2013). Postgenomics diagnostics: metabolomics approaches to human blood profiling. OMICS: A Journal of Integrative Biology, 17(11), 550–559. doi:10.1089/omi.2012.0121.
van den Berg, R. A., Hoefsloot, H. C. J., Westerhuis, J. A., Smilde, A. K., & van der Werf, M. J. (2006). Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC genomics, 7, 142. doi:10.1186/1471-2164-7-142.
Wang, S., Kuo, C., & Tseng, Y. J. (2013). Batch normalizer: A fast total abundance regression calibration method to simultaneouly adjust batch and injection order effects in liquid chromatography/time-of-fligth mass spectrometry-based metabolomics data and comparison with current calibration meth. Analytical Chemistry, 85, 1037–1046.
WHO. (2003). Screening for type 2 diabetes.
WHO. (1999) Definition, diagnosis and classification of diabetes mellitus and its complications.
Xia, J., Psychogios, N., Young, N., & Wishart, D. S. (2009). MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Research, 37, W652–W660. doi:10.1093/nar/gkp356.
Xu, F., Tavintharan, S., Sum, C. F., Woon, K., Lim, S. C., & Ong, C. N. (2013). Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. The Journal of Clinical Endocrinology and Metabolism, 98(6), E1060–E1065. doi:10.1210/jc.2012-4132.
Yagil, C., Barkalifa, R., Sapojnikov, M., Wechsler, A., Ben-Dor, D., Weksler-Zangen, S., et al. (2007). Metabolic and genomic dissection of diabetes in the Cohen rat. Physiological Genomics, 29(2), 181–192. doi:10.1152/physiolgenomics.00210.2006.
Zelena, E., Dunn, W. B., Broadhurst, D., Francis-McIntyre, S., Carroll, K. M., Begley, P., et al. (2009). Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Analytical Chemistry, 81(4), 1357–1364. doi:10.1021/ac8019366.
Zeng, M., Che, Z., Liang, Y., Wang, B., Chen, X., Li, H., et al. (2009). GC–MS based plasma metabolic profiling of type 2 diabetes mellitus. Chromatographia, 69(9–10), 941–948. doi:10.1365/s10337-009-1040-0.
Zhang, A. H., Qiu, S., Xu, H. Y., Sun, H., & Wang, X. J. (2014). Metabolomics in diabetes. Clinica Chimica Acta, 429, 106–110. doi:10.1016/j.cca.2013.11.037.
Acknowledgments
We acknowledge the Agilent Technologies Brasil Ltda, Life Sciences & Chemical Analysis for the use of the GC/MS system (7890B gas chromatograph coupled to a mass selective detector model 5977A-Agilent) for support data collection.
Funding information
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP 2012/05447-0, 2012/12042-7, 2013-17368-0, PROADI_ SUS Project nº 25000.180664/2011-35).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All experiments were carried out in accordance with the ethics committee of the Hospital das Clinicas, University of São Paulo, Brazil (protocol number 3759/12/015), approved the study protocol.
Informed consent
All participants signed a written informed consent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
PCA scores plot based on the GC/MS spectra of serum samples obtained from diabetic patients treated and diabetic patients not treated with hypoglycemic medication. (PDF 20 kb)
Figure S2
Samples first principal component scores distributed by the day of analysis. The red points represent the diabetic samples subject. (TIFF 1346 kb)
Figure S3
Metabolites concentration before (A) and after (B) LOESS normalization according to injection order and day of analysis. (TIFF 2194 kb)
Figure S4
Representativity of significant metabolites found by polygenic analysis. The blue side represent diabetic group, and the red side represent the non-diabetic group. (TIFF 1426 kb)
Table S1
Main biological functions disturbed relate with metabolites found by PLS-DA to diabetic, pre-diabetic and non diabetic participants. In #molecules column is the number of metabolites present in each pathway. (DOCX 15 kb)
Table S2
Heritability (h2) estimates (in percentage) for each metabolite, unadjusted (Model 1), adjusted for age and sex (Model 2), and adjusted for age, sex and T2DM (Model 3). (XLSX 15 kb)
Table S3
Main biological functions disturbed by proximal metabolites. In #molecules column is the number of metabolites present in each pathway. (DOCX 15 kb)
Table S4
Main biological functions related to distal metabolites. In #molecules column is the number of metabolites present in each pathway. (DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Padilha, K., Venturini, G., de Farias Pires, T. et al. Serum metabolomics profile of type 2 diabetes mellitus in a Brazilian rural population. Metabolomics 12, 156 (2016). https://doi.org/10.1007/s11306-016-1107-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-016-1107-5