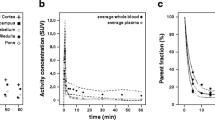
Abstract
Purpose
This study evaluates the performance of several parametric methods for assessing [11C]flumazenil binding distribution in the rat brain.
Procedures
Dynamic (60 min) positron emission tomography data with metabolite-corrected plasma input function were retrospectively analyzed (male Wistar rats, n = 10). Distribution volume (V T) images were generated from basis function method (BFM), Logan graphical analysis (Logan), and spectral analysis (SA). Using the pons as pseudo-reference tissue, binding potential (BP ND and DVR–1) images were obtained from receptor parametric imaging algorithms (RPM and SRTM2) and reference Logan (RLogan). Standardized uptake value images (SUV and SUVR) were also computed for different intervals post-injection. Next, regional averages were extracted from the parametric images, using pre-defined volumes of interest, which were also applied to the regional time-activity curves from the dynamic data. Parametric data were compared to their regional counterparts and to two-tissue compartment model (2TCM)-based values (previously defined as the model of choice for rats). Parameter agreement was assessed by linear regression analysis and Bland-Altman plots.
Results
All parametric methods strongly correlated to their regional counterparts (R 2 > 0.97) and to the 2TCM values (R 2 ≥ 0.95). SA and RLogan underestimated V T and BP ND (slope of 0.93 and 0.86, respectively), while SUVR-1 overestimated BP ND (slope higher than 1.07 for all intervals). While BFM and SRTM2 had the smallest bias to 2TCM values (0.05 for both), ratio Bland-Altman plots showed Logan and RLogan displayed relative errors which were comparable between different regions, in contrast with the other methods. Although SUV consistently underestimated V T, the bias in this method was also constant across regions.
Conclusions
All parametric methods performed well for the analysis of [11C]flumazenil distribution and binding in the rat brain. However, Logan and RLogan slightly outperformed the other methods in terms of precision, providing robust parameter estimation and constant bias. Yet, other methods can be of interest, because they can provide tissue perfusion (i.e., K 1 with BFM and SA), relative flow (i.e., R 1 with RPM and SRTM2), and model order (SA) images.
Similar content being viewed by others
Introduction
The in vivo study of neuronal integrity has been well established by means of positron emission tomography (PET) imaging with [11C]flumazenil [1, 2]. A number of different conditions have previously been studied with [11C]flumazenil [3,4,5], and the link between the GABA-ergic system and neurological disorders [6] and inflammation [7] has increased the applicability and interest in PET imaging with this radiotracer.
In addition to the non-invasive character of PET, one of its main advantages for studying physiological processes is that it can provide quantitative information. Full quantitative analysis can be achieved by pharmacokinetic modeling, which describes the time course of the PET tracer in tissue and enables the estimation of different parameters related to tracer distribution, metabolism, or receptor density and binding [8, 9]. In that context, the quantification of [11C]flumazenil binding has been previously performed by different pharmacokinetic models. In human studies, the one-tissue compartment model (1TCM) and the simplified reference tissue model (SRTM) [10] using the pons as a reference tissue have been validated [11] and applied [12,13,14]. Although useful, these models have been mostly applied for regional quantification of [11C]flumazenil binding, analyzing the average kinetic profile of a specific volume of interest (VOI).
In cases when subtle or disease-specific changes are expected, VOI-based pharmacokinetic modeling might be suboptimal precisely due to the use of pre-defined VOIs. If physiological changes are restricted to a subset of a region (tissue heterogeneity) or if they do not follow anatomical delineations, the average signal from pre-defined VOIs might not be able to properly describe the underlying alterations. There, the generation and analysis of parametric images can be of greater use. Parametric images are graphical representations of quantitative endpoints, where every image voxel corresponds to a kinetic parameter, such as distribution volume (V T) or binding potential (BP ND). Although parametric imaging is most commonly applied to human studies, it could be of great interest for the analysis of animal PET data. Preclinical PET imaging plays an important role in drug development and treatment assessment studies, for example, and it could be advantageous to explore full kinetic analysis of subtle physiological changes by parametric imaging also in the context of such animal study designs. Moreover, parametric imaging enables group comparisons at the voxel level using statistical parametric mapping (SPM), which performs the statistical analysis independent of pre-defined regions of interest.
In human studies, parametric images of [11C]flumazenil binding have been generated from a number of different methods, and the Logan graphical analysis, a multilinear reference tissue model (MRTM2), and the receptor parametric mapping method (RPM) were found to provide the best quantitative metrics [15]. However, no similar analysis has been performed for animal data. Moreover, the preference and performance of VOI-based models have already been shown to differ between human and rat studies [16]. Together with the higher noise levels typical of animal PET data, this difference could indicate the parametric models found to work best for the analysis of human studies might not be the most appropriate in the pre-clinical setting.
The aim of the current study was, therefore, to investigate the performance of several parametric methods for the analysis of [11C]flumazenil PET images of the rat brain. To that end, we retrospectively analyzed [11C]flumazenil pre-clinical images with both plasma input and reference tissue-based parametric methods and compared the results to their corresponding VOI-based pharmacokinetic models.
Material and Methods
Animal Data
We retrospectively analyzed male Wistar-Unilever outbred rats (n = 10) obtained from Harlan (Horst, The Netherlands). The animal experiments were performed according to the Dutch Law on Animal Experiments and were approved by the Institutional Animal Care and Use Committee of the University of Groningen (6264B).
Data Acquisition
Prior to the PET scan, a mixture of 5 % isoflurane and medical air was used to anesthetize the animals, which were maintained under anesthesia at 1.5–2.0 %, with a flow of 1.5–2 ml/min. Scans were performed in the Focus 220 camera (Siemens Medical Solutions, USA), where animals were positioned transaxially with the head in the field of view. First, a transmission scan was performed with a point-source of Co-57. Next, [11C]flumazenil was injected (bolus injection) with an automatic pump over 60s, and the PET camera was started at the moment of injection. List-mode PET data was acquired for a period of 60 min. [11C]Flumazenil was synthesized as previously described [17], and a 53 ± 18 % radiochemical yield was obtained, with radiochemical purity of [11C]flumazenil of 100 % and a pH of 6.5–7. The mean injected activity was 59.3 ± 22.6 MBq, with a 3.3 ± 2.4 nmol of injected mass, and the weight of the animals was 0.273 ± 0.03 kg [18].
During the scan, arterial blood samples (0.1 ml) were obtained (n = 16) for each individual animal, corresponding to 5, 10, 15, 30, 45, and 60 min post-injection (p.i.). The samples were subsequently separated into blood and plasma, and the activity was measured in a gamma counter (LKB-Wallac, Finland). For the metabolite analysis, 2–3 larger samples (0.6 ml) were collected for each animal, and a population metabolite curve was constructed by combining data from the individual samples. The validity of using a population-based metabolite curve in this animal group was previously assessed and reported elsewhere [17, 18]. Finally, this curve was used for the construction of individual metabolite-corrected plasma input functions.
Image Processing
After all necessary corrections, the list-mode data was reconstructed into 21 frames (6 × 10, 4 × 30, 2 × 60, 1 × 120, 1 × 180, 4 × 300, and 3 × 600 s). The 2D–OSEM algorithm was used to reconstruct the Fourier rebinned sinograms, with 4 iterations and 16 subsets. The resulting images displayed a 128 × 128 × 95 matrix, 0.63 mm pixel width, and 0.79 mm slice thickness. Next, individual dynamic images were coregistered to a [11C]flumazenil template [19] in PMOD v3.7 (PMOD Technologies Ltd., Switzerland). Time-activity curves (TACs) were then generated from a set of pre-defined bilateral VOIs which included the whole brain, regions with high GABAA expression (frontal cortex and hippocampus), and regions with low GABAA expression (cerebellum, medulla, and pons). Following previous studies [16], the pons was set as the pseudo-reference tissue and its TAC was used as input for reference-based models. In order to minimize bias induced by noise in the parametric methods, an isotropic 3D Gaussian filter (FWHM = 0.8 mm) was applied. Finally, individually generated brain masks were also applied prior to modeling.
Pharmacokinetic Modeling
All pharmacokinetic modeling was performed with the PPET [20] software package.
First, parametric V T images were generated from three different methods, including the basis function method (BFM), which is an implementation of the 1TCM, the Logan graphical analysis (Logan) [21], and the spectral analysis (SA) [22]. For these methods, the metabolite-corrected plasma curve was used as input function, and blood volume fraction was accounted for as a fit parameter in BFM and SA. Next, parametric BP ND images were computed from three reference-based models. The first was the reference tissue Logan graphical analysis (RLogan) [23], which provides the distribution volume ratio (DVR) of target regions relative to the reference tissue. Following the relationship BP ND = DVR−1 [8], we subtracted 1 from the RLogan DVR to allow direct comparison to parametric images of BP ND. The other two methods consisted of two implementations of the receptor parametric mapping method (RPM and SRTM2) [24], which correspond to the two versions of the simplified reference tissue model (SRTM and SRTM2). For the SRTM2, the model fit was performed in two steps, where the median value of the reference tissue efflux rate constant (\( {k}_2^{\prime } \)) obtained from all voxels outside the reference tissue in the first run was entered as a fixed parameter for the second run [25]. An overview of the settings used for the parametric methods and optimized for this group can be found in Table 1.
Parametric images of standardized uptake value (SUV) and SUV target to pons ratio (SUVR) were also generated for the intervals 30–40 min, 40–50 min, and 50–60 min post-injection and subsequently referred to by SUVstart-end and SUVRstart-end. In order to compare SUVR and BP ND images, 1 was subtracted from the derived SUVR values, assuming equilibrium was reached for those intervals.
Method Evaluation
Results from each of the parametric methods were compared to (1) the corresponding regional (VOI) analysis using equivalent kinetic models but also to (2) reference values based on regional two-tissue compartment model (2TCM) fits. The 2TCM-derived parameters (V T and DVR−1) were considered as reference values for method comparison. An overview of parametric methods, their corresponding VOI-models, parameter of interest, and equivalent 2TCM parameter can be found in Table 2.
For that purpose, first the same pre-defined set of VOIs used for the generation of TACs were projected onto the parametric images, and average parameter estimates were obtained for each region. Next, the regional TACs were analyzed by six models corresponding to the parametric methods (i.e., 1TCM, Logan, SA, RLogan, SRTM, and SRTM2) and by the 2TCM. In the plasma input models, blood- and metabolite-corrected plasma curves were used as input function, with a fixed blood volume of 5 %. Finally, the regionally averaged values from parametric methods were compared to both their VOI-counterparts and to the corresponding 2TCM-derived reference.
Statistical Analysis
Parameter agreement was assessed by linear regression analysis and Bland-Altman plots. Results were considered significant when p < 0.05 and are expressed as mean ± SD.
Results
Volume of Distribution
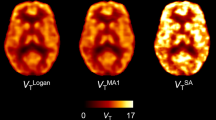
Visual inspection of parametric V T images showed good correspondence between methods and the expected [11C]flumazenil distribution in the rat brain (Fig. 1).
Compared to their VOI-counterpart, parametric Logan was the method with the best correlation, displaying an R 2 = 0.99 and a slope of 0.97 when the regression line was set through the origin (Table 3). The BFM also strongly correlated to its counterpart (1TCM) values (R 2 = 0.98), with a slope of 1.03. The slope of the linear regression of SA with its counterpart was further from identity (0.89), and the correlation was somewhat lower than the other methods (R 2 = 0.96). Overall, all three methods showed excellent correlation (R 2 = 0.99) to 2TCM reference V T values (Fig. 2a). However, BFM was the method which displayed the slope closest to the identity line (0.98). Although SA showed the same correlation coefficient as the other two methods, a slope of 0.93 indicated a larger underestimation in V T values.
Regression analysis and Bland-Altman plots of V T compared to 2TCM values. a Linear regression between parametric (BFM, Logan, and SA) and 2TCM V T values determined by VOI analysis. The solid line is the identity line, and the dashed lines represent the regression lines. b Bland-Altman plot of the agreement in V T estimation between the parametric methods and the reference values obtained from the 2TCM (VOI). The dashed line represents zero bias. c Ratio Bland-Altman plot between BFM, Logan, SA, and 2TCM V T values. The dashed line represents a ratio of one, corresponding to full agreement between methods.
The Bland-Altman analysis showed that, despite the good correlation, a negative bias (−0.20) was present with the Logan method (Fig. 2b). The BFM had the smallest overall bias in relation to 2TCM V T (0.05), yet its 95 % limits of agreement were slightly wider than those of Logan (Table 4). SA had the widest 95 % limits of agreement similar to BFM and an overall higher (negative) bias (−0.49). Figure 2c shows a ratio Bland-Altman plot to highlight the performance in parameter agreement for the different levels of V T values. There, regions with high V T (≥6) had a ratio close to one in comparison with 2TCM V T values. On the other hand, larger over- and underestimations occurred for regions of lower V T (<6).
Figure 3 displays the range of differences to 2TCM values for the three parametric methods across all regions. Significant differences in bias were found in all three methods when compared to 2TCM values. However, the bias from parametric Logan compared to 2TCM values showed the smallest variability (SD = 0.19), indicating higher precision for this method compared to the others.
Binding Potential (BP ND and DVR−1)
As can be seen in Fig. 1, the highest [11C]flumazenil binding spatially matched regions with a high receptor density (cortical and sub-cortical areas), while the lowest was seen in low-density ones (cerebellum and brainstem). Images of receptor binding were also visually comparable between parametric methods.
All reference-based methods performed well in comparison to 2TCM DVR−1 (Fig. 4a). In particular, DVR–1 from RLogan displayed the best correlation coefficient in relation to 2TCM values (R 2 = 0.99), while the slope was the furthest from the identity line (0.86), indicating underestimation of receptor binding. Binding estimates from RPM showed correlation to 2TCM values similar to those of RLogan (R 2 = 0.98) and a slope closer to identity (0.93) (Table 3). Estimation of binding from parametric SRTM2 outperformed both other methods in terms of linear regression, with a R 2 of 0.98 and a slope of 0.98 compared to 2TCM values.
Regression analysis and Bland-Altman plots of BP ND. a Linear regression between parametric (RLogan, RPM, and SRTM2) and 2TCM DVR–1 (BP ND) values determined by VOI analysis. The solid line represents the identity line, and the dashed lines correspond to each of the regression lines. b Bland-Altman plot of the agreement in BP ND estimation between the parametric methods and the reference values obtained from the 2TCM (VOI). The dashed line corresponds to zero bias. c Ratio Bland-Altman plot between BP ND of parametric methods and 2TCM BP ND. The dashed line corresponds to a ratio of one, representing full agreement between methods. The y-axis is truncated at −1.0.
Bland-Altman plots indicated a similar performance between RLogan and RPM methods (Fig. 4b). Both parametric methods displayed a negative bias compared to 2TCM DVR–1 (−0.17 for RLogan and −0.10 for RPM). SRTM2 showed a small overall bias (0.05) but resulted in the largest 95 % limits of agreement to 2TCM DVR–1, as can be seen in Table 4. For most levels of specific binding, all three methods showed good relative agreement, with a ratio close to one in comparison with 2TCM DVR–1 values (Fig. 4c). However, relative errors were larger for BP ND values close to zero.
In Fig. 3, a similar performance between the reference methods can be seen. Significant differences were found in the bias of parametric RLogan and SRTM2 when compared to 2TCM values. No significant difference was seen for the RPM bias to 2TCM values (p = 0.20). Yet, RLogan displayed the highest precision (SD = 0.72) compared to the other two methods.
SUV and SUVR–1
Correlation between parametric SUV and 2TCM V T was overall strong, with similar correlation coefficients across intervals p.i. (R 2 = 0.96, R 2 = 0.96, and R 2 = 0.95 for SUV30–40, SUV40–50, and SUV50–60, respectively). However, there was a clear scale difference between the two parameters, and a direct comparison was not possible, as SUV ranged from 0.2 to 2.7 and 2TCM V T ranged from 1.9 to 9.9 (Fig. 5a). However, ratio Bland-Altman plots showed that SUV from all three intervals underestimated 2TCM V T in a similar manner across regions, independent of the levels of uptake (Fig. 5b).
Regression analysis and ratio Bland-Altman plot of SUV from different intervals. a Regression analysis between SUV (parametric) and 2TCM V T (VOI) for three different intervals p.i. The dashed lines represent the regression lines. b Ratio Bland-Altman plot between V T estimates from SUV and from 2TCM V T (VOI) for the same intervals.
SUVR–1 images showed excellent correlation to binding estimates from 2TCM for all intervals p.i. (Fig. 6a). However, an overestimation of binding potential from SUVR–1 images was seen, with linear regression (through origin) slopes of 1.13, 1.21, and 1.07 for the SUVR30–40–1, SUVR40–50–1, and SUVR50–60–1, respectively. This was confirmed by a Bland-Altman analysis, where SUVR40–50–1 showed the highest bias compared to 2TCM DVR–1 and the largest 95 % limits of agreement (Table 4). In a ratio Bland-Altman, all intervals show similar performance, with the ratio close to one for most levels of specific binding (Fig. 6b). For binding values close to zero, relative errors were more pronounced, independent of the SUVR interval.
Regression analysis and ratio Bland-Altman plot of SUVR from different intervals. a Regression analysis between SUVR–1 (parametric) and 2TCM BP ND (VOI) for three different p.i. intervals. The solid line represents the identity line, and the dashed lines correspond to each of the regression lines. b Ratio Bland-Altman plot between binding potential estimates from SUVR–1 and from 2TCM BP ND (VOI) for the same intervals. The dashed line corresponds to a ratio of one, representing full agreement between methods. The y-axis is truncated at −1.0.
Parametric Images of K 1, R 1, and Model Order
For BFM and SA, parametric images of K 1 (influx rate constant) were also available (Fig. 1). Although both displayed similar perfusion, BFM K 1 was generally higher, while the less homogeneous distribution seen in the SA image suggested higher noise sensitivity for that method. SA also provided parametric images of model order, displaying the difference in the complexity of kinetic profiles between different voxels (Fig. 7). In those, a marked difference between high- and low-density regions is seen, and a more complex model was more frequently observed in the later, compared to the former.
Additional parametric images were also available from RPM and SRTM2 displaying R 1, i.e., the ratio between K 1 of a target region and of the pseudo-reference tissue (pons). As can be seen in Fig. 1, there is a good correspondence between the two methods. However, R 1 images from SRTM2 displayed better contrast and lower noise levels compared to RPM R 1 images.
Significant (p < 0.01) within-animal correlations were found between K 1 and R 1 values of parametric and VOI-counterparts for all animals when pooling the models (Spearman’s rho ranged from 0.57 to 0.80). However, when comparing parametric values to reference 2TCM estimates, significant within-animal correlations were found for only 7 out of 10 animals. Moreover, by grouping the animals and comparing estimates by model, only BFM K 1 values were significantly correlated to VOI-counterpart estimates (Spearman’s rho 0.68, p < 0.01).
Discussion
The use of parametric imaging methods for the analysis of PET data can be of importance when subtle and/or disease-specific changes are expected. In the case of [11C]flumazenil brain PET, those changes are related to the GABA-ergic system and can be present in several conditions related to neuronal loss and inflammatory processes. While parametric images of [11C]flumazenil distribution and binding have been generated and applied in human studies, the same has not yet been done for the pre-clinical setting. Therefore, this study evaluated several parametric methods for the visualization and analysis of [11C]flumazenil PET images of the rat brain.
A first analysis of parametric V T images demonstrated that all three methods tested in this study (BFM, Logan, and SA) corresponded well to their VOI-counterparts. These results indicate that the generation of parametric V T maps from these methods could, to some extent, replicate results obtained from a regional analysis, which is generally less affected by noise and image resolution. More importantly, there was also an excellent correlation between parametric values of each of the three parametric methods and V T estimated from the reference 2TCM VOI analysis. In terms of parameter agreement, however, a slight difference between the methods becomes noticeable. While BFM V T showed a small bias compared to 2TCM values (0.05), the wide 95 % limits of agreement suggest this method might not be optimal for parameter estimation at the individual level. A similar behavior could be seen with V T estimates from SA. This variability was especially true for regions of small V T (low-density regions). This observation is in agreement with a previous report of a VOI analysis, where we found the difference between 1TCM and 2TCM V T values to be larger for cerebellum, medulla, and pons [16]. Indeed, model differences seemed to be a function of receptor density and consequently, a function of the underlying tissue configuration. As shown in our previous study [18], a 1TCM configuration leads to major errors in parameter estimation in low-binding regions, both from plasma input models and reference models relying on fast (1 T) kinetics for the reference tissue. On the other hand, such a range of differences might not be relevant for most study designs, as regions with low-density of receptors are generally not the main focus of an analysis. Nonetheless, even though this region-dependent range of agreement was seen in all three methods, the range of differences to 2TCM values was smallest for Logan, which does not assume a specific compartmental configuration (Fig. 3). As a consequence, despite the significant bias compared to reference values, Logan can be considered the most robust of the three methods for the generation of parametric V T images, showing a higher precision independent of receptor density levels. Yet, it is important to note that while VOI analyses were performed with a fixed blood volume of 5 %, parametric methods either used it as a fit parameter or freely incorporated its contribution into the model. Although a limitation, previous work has shown that different contributions of blood to the overall signal did not significantly affect the main outcome parameter [18].
Parametric images of receptor binding were also comparable between different parametric methods. Moreover, a good correspondence to VOI-counterparts was seen for all reference methods, with high R 2 values and only small over- and underestimations seen for RPM and SRTM2, respectively (Table 3). In comparison with reference 2TCM values, correlation remained high for all methods, and linear regression slopes were closer to the identity line for the SRTM2. However, RLogan consistently underestimated binding, with a regression slope of 0.86. Interestingly, while RPM and SRTM2 had overall bias similar to those of RLogan (−0.10, 0.05, and −0.17, respectively), the range of differences to 2TCM values was larger for those methods (Fig. 3). In fact, the differences seen between SRTM2 and 2TCM estimates reached 0.5 for regions of low-binding—values higher than the actual BP ND estimates—translating into relative errors of more than 100 % (Fig. 4b, c). However, those correspond to BP ND values close to zero, where large relative errors are not surprising, nor meaningful. On the other hand, it is also important to notice that all reference-based methods underestimate the true BP ND from the start, due to the considerable levels of specific binding present in the rat pons [16]. Nonetheless, RLogan displayed the highest precision in estimating receptor binding, as can be seen in Fig. 3. Therefore, despite its bias to 2TCM values, RLogan can be considered the most robust reference-based parametric method for the generation of receptor binding images.
While V T and BP ND are important kinetic parameters, both require dynamic scanning for their estimation. As a consequence, SUV and SUVR determined from static late images are widely applied for both regional and parametric analyses. While SUV does not necessarily correspond to V T, both parameters are directly related. This was also seen in our findings, where good correlation (R 2 > 0.95) was found between parametric SUV determined at three p.i. intervals and 2TCM V T (Fig. 5a). In turn, SUVR is closely related to BP ND, and when equilibrium is reached, SUVR–1 is expected to correspond to DVR–1. Therefore, a more direct comparison between SUVR–1 from different intervals p.i. and BP ND could be performed. However, as can be seen in Fig. 1, this method overestimated BP ND from other methods and DVR–1 from 2TCM (Table 3). The largest overestimation of 2TCM DVR–1 was seen for SUVR40–50 and the smallest for SUVR50–60, which might indicate equilibrium is not reached at earlier intervals. Nonetheless, all SUVR–1 strongly correlated to 2TCM DVR–1. It is important to notice that despite the high correlations, the performance of SUV and SUVR–1 was region-dependent (Figs. 5a and 6a, respectively). However, when assessing agreement by a ratio Bland-Altman, the results suggested a scaling effect for SUV, since relative errors were similar across regions (Fig. 5b). Although useful, SUV and SUVR images must be applied carefully in group comparisons, interventional and longitudinal studies, since they are especially affected by changes in metabolism and tissue perfusion [26].
The results of this study indicate that parametric analysis of [11C]flumazenil images of the rat brain can be satisfactorily performed, and, as a consequence, parametric imaging and SPM analysis would allow direct statistical comparison across groups and/or conditions without prior assumptions on regions of interest. In fact, SPM analysis has previously been able to detect more subtle disease-related changes in an animal model of inflammation imaged with [11C]PK11195, for example [27]. However, the homogeneous dataset analyzed in this study did not allow for a similar assessment of the performance of parametric imaging in group comparison nor was this within the scope of the study. Thus, further studies should be performed in order to evaluate the benefit of parametric imaging of [11C]flumazenil binding in the rat brain for different conditions and study designs. In general, since Logan and RLogan outperformed other methods in precision and are of robust and simple implementation, both methods are preferred for group comparisons and longitudinal studies. On the other hand, alternatives such as BFM or RPM can be of value in generating parametric images with additional information (K 1 or R 1), while SUV and SUVR images could be useful when dynamic scanning is not possible, despite under- and overestimation of kinetic parameters.
Conclusion
This study demonstrated several parametric methods performed well for the generation of [11C]flumazenil distribution and binding images, with a good correlation to 2TCM-derived values being observed across all methods. Therefore, a specific method should be applied according to what information is of interest to a specific research question. Nonetheless, the most precise and straightforward methods were found to be Logan and RLogan, while BFM or RPM could be considered when information on perfusion is of significance.
Change history
02 January 2018
The name of the second author was incorrectly cited and is corrected now, as displayed here.
References
Heiss WD, Grond M, Thiel A et al (1998) Permanent cortical damage detected by flumazenil positron emission tomography in acute stroke. Stroke 29:454–461
Heiss WD, Kracht L, Grond M et al (2000) Early [11C]flumazenil/H(2)O positron emission tomography predicts irreversible ischemic cortical damage in stroke patients receiving acute thrombolytic therapy. Stroke 31:366–369
Tian J, Yong J, Dang H, Kaufman DL (2011) Oral GABA treatment downregulates inflammatory responses in a mouse model of rheumatoid arthritis. Autoimmunity 44:465–470
Heiss WD, Sobesky J, Smekal U et al (2004) Probability of cortical infarction predicted by flumazenil binding and diffusion-weighted imaging signal intensity: a comparative positron emission tomography/magnetic resonance imaging study in early ischemic stroke. Stroke 35:1892–1898
Rojas S, Martín A, Pareto D et al (2011) Positron emission tomography with 11C-flumazenil in the rat shows preservation of binding sites during the acute phase after 2h-transient focal ischemia. Neuroscience 182:208–216
Wong CGT, Bottiglieri T, Snead OC (2003) GABA, gamma-hydroxybutyric acid, and neurological disease. Ann Neurol 54(Suppl 6):S3–12
Barragan A, Weidner JM, Jin Z et al (2015) GABAergic signalling in the immune system. Acta Physiol (Oxf) 213:819–827
Innis RB, Cunningham VJ, Delforge J et al (2007) Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab 27:1533–1539
Gunn RN, Gunn SR, Cunningham VJ (2001) Positron emission tomography compartmental models. J Cereb Blood Flow Metab 21:635–652
Lammertsma AA, Hume SP (1996) Simplified reference tissue model for PET receptor studies. NeuroImage 4:153–158
Koeppe RA, Holthoff VA, Frey KA et al (1991) Compartmental analysis of [11C]flumazenil kinetics for the estimation of ligand transport rate and receptor distribution using positron emission tomography. J Cereb Blood Flow Metab 11:735–744
Geeraerts T, Coles JP, Aigbirhio FI et al (2011) Validation of reference tissue modelling for [11C]flumazenil positron emission tomography following head injury. Ann Nucl Med 25:396–405
Klumpers UM, Veltman DJ, Boellaard R et al (2008) Comparison of plasma input and reference tissue models for analysing [11C]flumazenil studies. J Cereb Blood Flow Metab 28:579–587
Salmi E, Aalto S, Hirvonen J et al (2008) Measurement of GABAA receptor binding in vivo with [11C]flumazenil: a test-retest study in healthy subjects. NeuroImage 41:260–269
Klumpers UMH, Boellaard R, Veltman DJ et al (2012) Parametric [11C]flumazenil images. Nucl Med Commun 33:422–430
Lopes Alves I, Vállez García D, Parente A, et al. (2016) [11C]flumazenil kinetics in the rat brain: model preference and the impact of non-specific and non-selective binding in reference region modeling. Eur J Nucl Med Mol Imaging 43:S25.
Parente A, García DV, Shoji A et al (2017) Contribution of neuroinflammation to changes in [11C]flumazenil binding in the rat brain: evaluation of the inflamed pons as reference tissue. Nucl Med Biol 49:50–56
Alves IL, Vállez García D, Parente A et al (2017) Pharmacokinetic modeling of [11C]flumazenil kinetics in the rat brain. EJNMMI Res 7:17
Vállez Garcia D, Casteels C, Schwarz AJ et al (2015) A standardized method for the construction of tracer specific PET and SPECT rat brain templates: validation and implementation of a toolbox. PLoS One 10:e0122363
Boellaard R, Yaqub M, Lubberink M, Lammertsma A (2006) PPET: a software tool for kinetic and parametric analyses of dynamic PET studies. NeuroImage 31:T62
Logan J, Fowler JS, Volkow ND et al (1990) Graphical analysis of reversible radioligand binding from time—activity measurements applied to [N-11C-methyl]-(−)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab 10:740–747
Cunningham VJ, Jones T (1993) Spectral analysis of dynamic PET studies. J Cereb Blood Flow Metab 13:15–23
Logan J, Fowler JS, Volkow ND et al (1996) Distribution volume ratios without blood sampling from graphical analysis of PET data. J Cereb Blood Flow Metab 16:834–840
Gunn RN, Lammertsma AA, Hume SP, Cunningham VJ (1997) Parametric imaging of ligand-receptor binding in PET using a simplified reference region model. NeuroImage 6:279–287
Wu Y, Carson RE (2002) Noise reduction in the simplified reference tissue model for neuroreceptor functional imaging. J Cereb Blood Flow Metab 22:1440–1452
van Berckel BNM, Ossenkoppele R, Tolboom N et al (2013) Longitudinal amyloid imaging using 11C-PiB: methodologic considerations. J Nucl Med 54:1570–1576
Parente A, Feltes PK, Vallez Garcia D et al (2016) Pharmacokinetic analysis of 11C-PBR28 in the rat model of herpes encephalitis: comparison with (R)- 11C-PK11195. J Nucl Med 57:785–791
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The scholarship of Andrea Parente was financed by Siemens. The other authors declare no conflict of interest.
Additional information
A correction to this article is available online at https://doi.org/10.1007/s11307-017-1156-9.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lopes Alves, I., Vállez García, D.V., Parente, A. et al. Parametric Imaging of [11C]Flumazenil Binding in the Rat Brain. Mol Imaging Biol 20, 114–123 (2018). https://doi.org/10.1007/s11307-017-1098-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-017-1098-2