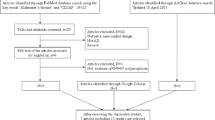
Abstract
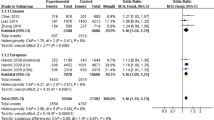
The CD33 rs3865444 polymorphism was first identified to be associated with Alzheimer’s disease (AD) in European population. However, the following studies reported weak or no significant association in Chinese, Japanese, Korean, American, and Canadian populations. We think that these negative results may have been caused by either relatively small sample sizes compared with those used for the previous GWAS in European ancestry or the genetic heterogeneity of the rs3865444 polymorphism in different populations. Here, we reevaluated this association using the relatively large-scale samples from previous 27 studies (N = 86,759; 31,106 cases and 55,653 controls) by searching the PubMed, AlzGene, and Google Scholar databases. We identified significant heterogeneity and observed no significant association between the rs3865444 polymorphism and AD in pooled populations (P = 0.264, odds ratio (OR) = 0.97, 95 % confidence interval (CI) 0.93–1.02). In subgroup analysis, we identified significant heterogeneity only in East Asian population and observed no significant association between the rs3865444 polymorphism and AD. We further identified significant heterogeneity and observed significant association between the rs3865444 polymorphism and AD in Chinese population. We identified no significant heterogeneity and significant association in North American and European populations. Collectively, our analysis shows that the CD33 rs3865444 polymorphism is associated with AD susceptibility in Chinese, European, and North American populations. We believe that our findings will be very useful for future genetic studies on AD.



Similar content being viewed by others
References
Liu G, Jiang Y, Wang P, Feng R, Jiang N, Chen X, Song H, Chen Z (2012) Cell adhesion molecules contribute to Alzheimer’s disease: multiple pathway analyses of two genome-wide association studies. J Neurochem 120:190–198
Liu G, Yao L, Liu J, Jiang Y, Ma G, Chen Z, Zhao B, Li K (2014) Cardiovascular disease contributes to Alzheimer’s disease: evidence from large-scale genome-wide association studies. Neurobiol Aging 35:786–792
Liu G, Wang H, Liu J, Li J, Li H, Ma G, Jiang Y, Chen Z, Zhao B, Li K (2014) The clu gene rs11136000 variant is significantly associated with Alzheimer’s disease in Caucasian and Asian populations. Neuromol Med 16:52–60
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD (2011) Common variants at ms4a4/ms4a6e, cd2ap, cd33 and epha1 are associated with late-onset Alzheimer’s disease. Nat Genet 43:436–441
Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J (2011) Common variants at abca7, ms4a6a/ms4a4e, epha1, cd33 and cd2ap are associated with Alzheimer’s disease. Nat Genet 43:429–435
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, Destefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45:1452–1458
Deng YL, Liu LH, Wang Y, Tang HD, Ren RJ, Xu W, Ma JF, Wang LL, Zhuang JP, Wang G, Chen SD (2012) The prevalence of cd33 and ms4a6a variant in Chinese Han population with Alzheimer’s disease. Hum Genet 131:1245–1249
Tan L, Yu JT, Zhang W, Wu ZC, Zhang Q, Liu QY, Wang W, Wang HF, Ma XY, Cui WZ (2013) Association of GWAS-linked loci with late-onset Alzheimer’s disease in a northern Han Chinese population. Alzheimers Dement 9:546–553
Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, Kawarabayashi T, Shoji M, Tomita N, Arai H, Asada T, Harigaya Y, Ikeda M, Amari M, Hanyu H, Higuchi S, Ikeuchi T, Nishizawa M, Suga M, Kawase Y, Akatsu H, Kosaka K, Yamamoto T, Imagawa M, Hamaguchi T, Yamada M, Moriaha T, Takeda M, Takao T, Nakata K, Fujisawa Y, Sasaki K, Watanabe K, Nakashima K, Urakami K, Ooya T, Takahashi M, Yuzuriha T, Serikawa K, Yoshimoto S, Nakagawa R, Kim JW, Ki CS, Won HH, Na DL, Seo SW, Mook-Jung I, St George-Hyslop P, Mayeux R, Haines JL, Pericak-Vance MA, Yoshida M, Nishida N, Tokunaga K, Yamamoto K, Tsuji S, Kanazawa I, Ihara Y, Schellenberg GD, Farrer LA, Kuwano R (2013) Sorl1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One 8:e58618
Chung SJ, Lee JH, Kim SY, You S, Kim MJ, Lee JY, Koh J (2013) Association of GWAS top hits with late-onset Alzheimer disease in Korean population. Alzheimer Dis Assoc Disord 27:250–257
Omoumi A, Fok A, Greenwood T, Sadovnick AD, Feldman HH, Hsiung GY (2014) Evaluation of late-onset Alzheimer disease genetic susceptibility risks in a Canadian population. Neurobiol Aging 35:936, e935-912
Logue MW, Schu M, Vardarajan BN, Buros J, Green RC, Go RC, Griffith P, Obisesan TO, Shatz R, Borenstein A, Cupples LA, Lunetta KL, Fallin MD, Baldwin CT, Farrer LA (2011) A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol 68:1569–1579
Lewis CM, Knight J (2012) Introduction to genetic association studies. Cold Spring Harb Protoc 2012:297–306
Liu G, Zhang S, Cai Z, Ma G, Zhang L, Jiang Y, Feng R, Liao M, Chen Z, Zhao B, Li K (2013) Picalm gene rs3851179 polymorphism contributes to Alzheimer’s disease in an Asian population. Neuromol Med 15:384–388
Jiang Y, Zhang R, Zheng J, Liu P, Tang G, Lv H, Zhang L, Shang Z, Zhan Y, Lv W, Shi M (2012) Meta-analysis of 125 rheumatoid arthritis-related single nucleotide polymorphisms studied in the past two decades. PLoS One 7:e51571
Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, Passmore P, Morgan K, Younkin SG (2011) Replication of epha1 and cd33 associations with late-onset Alzheimer’s disease: a multi-centre case–control study. Mol Neurodegener 6:54
Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von Korff A, Morris MC, Evans DA, Johnson K, Sperling RA, Schneider JA, Bennett DA, De Jager PL (2013) Cd33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci 16:848–850
Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE (2013) Alzheimer’s disease risk gene cd33 inhibits microglial uptake of amyloid beta. Neuron 78:631–643
Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A, Rothamel K, Stranger BE, Bennett DA, Evans DA, De Jager PL, Bradshaw EM (2014) Cd33: increased inclusion of exon 2 implicates the ig v-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet 23:2729–2736
Liu G, Zhang S, Cai Z, Li Y, Cui L, Ma G, Jiang Y, Zhang L, Feng R, Liao M, Chen Z, Zhao B, Li K (2013) Bin1 gene rs744373 polymorphism contributes to Alzheimer’s disease in East Asian population. Neurosci Lett 544:47–51
Liu G, Zhang L, Feng R, Liao M, Jiang Y, Chen Z, Zhao B, Li K (2013) Lack of association between picalm rs3851179 polymorphism and Alzheimer’s disease in Chinese population and apoeepsilon4-negative subgroup. Neurobiol Aging 34:1310, e1319-1310
Liu G, Li F, Zhang S, Jiang Y, Ma G, Shang H, Liu J, Feng R, Zhang L, Liao M, Zhao B, Li K (2014) Analyzing large-scale samples confirms the association between the abca7 rs3764650 polymorphism and Alzheimer’s disease susceptibility. Mol Neurobiol
Shen N, Chen B, Jiang Y, Feng R, Liao M, Zhang L, Li F, Ma G, Chen Z, Zhao B, Li K, Liu G (2014) An updated analysis with 85,939 samples confirms the association between cr1 rs6656401 polymorphism and Alzheimer’s disease. Mol Neurobiol
Zhang S, Zhang D, Jiang Y, Wu L, Shang H, Liu J, Feng R, Liao M, Zhang L, Liu Y, Liu G, Li K (2014) Clu rs2279590 polymorphism contributes to Alzheimer’s disease susceptibility in Caucasian and Asian populations. J Neural Transm
Gogele M, Minelli C, Thakkinstian A, Yurkiewich A, Pattaro C, Pramstaller PP, Little J, Attia J, Thompson JR (2012) Methods for meta-analyses of genome-wide association studies: critical assessment of empirical evidence. Am J Epidemiol 175:739–749
Acknowledgments
This work was supported by funding from the National Nature Science Foundation of China (grant numbers 81300945, 31200934, 31301938, 81471294, 31171219, 81271213, 81271214) and the Natural Science Foundation of Zhejiang Province, China (LY12H04006 to Li XW).
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xingwang Li, Ning Shen, and Shuyan Zhang contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 391 kb)
Rights and permissions
About this article
Cite this article
Li, X., Shen, N., Zhang, S. et al. CD33 rs3865444 Polymorphism Contributes to Alzheimer’s Disease Susceptibility in Chinese, European, and North American Populations. Mol Neurobiol 52, 414–421 (2015). https://doi.org/10.1007/s12035-014-8880-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-8880-9