Abstract
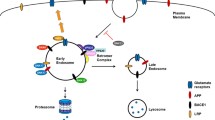
The amyloid precursor protein (APP), one key player in Alzheimer’s disease (AD), is extensively processed by different proteases. This leads to the generation of diverging fragments including the amyloid β (Aβ) peptide, which accumulates in brains of AD patients. Subcellular trafficking of APP is an important aspect for its proteolytic conversion, since the various secretases which cleave APP are located in different cellular compartments. As a consequence, altered subcellular targeting of APP is thought to directly affect the degree to which Aβ is generated. The mechanisms underlying intracellular APP transport are critical to understand AD pathogenesis and can serve as a target for future pharmacological interventions. In the recent years, a number of APP interacting proteins were identified which are implicated in sorting of APP, thereby influencing APP processing at different angles of the secretory or endocytic pathway. This review provides an update on the proteolytic processing of APP and the interplay of the transmembrane proteins low-density lipoprotein receptor-related protein 1, sortilin-receptor with A-type repeats, SorCS1c, sortilin, and calsyntenin. We discuss the specific interactions with APP, the capacity to modulate the intracellular itinerary and the proteolytic conversion of APP, a possible involvement in the clearance of Aβ, and the implications of these transmembrane proteins in AD and other neurodegenerative diseases.








Similar content being viewed by others
References
Huang Y, Mucke L (2012) Alzheimer mechanisms and therapeutic strategies. Cell 148(6):1204–1222
Tanzi RE, Bertram L (2005) Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell 120(4):545–555
Small SA, Gandy S (2006) Sorting through the cell biology of Alzheimer’s disease: intracellular pathways to pathogenesis. Neuron 52(1):15–31
King GD, Scott Turner R (2004) Adaptor protein interactions: modulators of amyloid precursor protein metabolism and Alzheimer’s disease risk? Exp Neurol 185(2):208–219
Back S et al (2007) Beta-amyloid precursor protein can be transported independent of any sorting signal to the axonal and dendritic compartment. J Neurosci Res 85(12):2580–2590
Szodorai A et al (2009) APP anterograde transport requires Rab3A GTPase activity for assembly of the transport vesicle. J Neurosci: Off J Soc Neurosci 29(46):14534–14544
Kuhn PH et al (2010) ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J 29(17):3020–3032
Lammich S et al (1999) Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A 96(7):3922–3927
Slack BE, Ma LK, Seah CC (2001) Constitutive shedding of the amyloid precursor protein ectodomain is up-regulated by tumour necrosis factor-alpha converting enzyme. Biochem J 357(Pt 3):787–794
Lichtenthaler SF (2011) Alpha-secretase in Alzheimer’s disease: molecular identity, regulation and therapeutic potential. J Neurochem 116(1):10–21
Lichtenthaler SF, Haass C, Steiner H (2011) Regulated intramembrane proteolysis—lessons from amyloid precursor protein processing. J Neurochem 117(5):779–796
Zhao G et al (2004) Identification of a new presenilin-dependent zeta-cleavage site within the transmembrane domain of amyloid precursor protein. J Biol Chem 279(49):50647–50650
Weidemann A et al (2002) A novel epsilon-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with notch processing. Biochemistry 41(8):2825–2835
Sastre M et al (2001) Presenilin-dependent gamma-secretase processing of beta-amyloid precursor protein at a site corresponding to the S3 cleavage of notch. EMBO Rep 2(9):835–841
Gu Y et al (2001) Distinct intramembrane cleavage of the beta-amyloid precursor protein family resembling gamma-secretase-like cleavage of notch. J Biol Chem 276(38):35235–35238
Haass C et al (1993) Beta-amyloid peptide and a 3-kDa fragment are derived by distinct cellular mechanisms. J Biol Chem 268(5):3021–3024
Vassar R et al (1999) Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286(5440):735–741
Vassar R et al (2009) The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J Neurosci 29(41):12787–12794
Haass C et al (1992) Amyloid beta-peptide is produced by cultured cells during normal metabolism. Nature 359(6393):322–325
Qi-Takahara Y et al (2005) Longer forms of amyloid beta protein: implications for the mechanism of intramembrane cleavage by gamma-secretase. J Neurosci Off J Soc Neurosci 25(2):436–445
Xia W et al (1997) Enhanced production and oligomerization of the 42-residue amyloid beta-protein by Chinese hamster ovary cells stably expressing mutant presenilins. J Biol Chem 272(12):7977–7982
Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297(5580):353–356
Selkoe DJ, Wolfe MS (2007) Presenilin: running with scissors in the membrane. Cell 131(2):215–221
Weggen S, Beher D (2012) Molecular consequences of amyloid precursor protein and presenilin mutations causing autosomal-dominant Alzheimer’s disease. Alzheimers Res Ther 4(2):9
Duering M et al (2005) Mean age of onset in familial Alzheimer’s disease is determined by amyloid beta 42. Neurobiol Aging 26(6):785–788
Takami M et al (2009) Gamma-secretase: successive tripeptide and tetrapeptide release from the transmembrane domain of beta-carboxyl terminal fragment. J Neurosci: Off J Soc Neurosci 29(41):13042–13052
Andrew RJ et al (2016) A Greek tragedy: the growing complexity of Alzheimer amyloid precursor protein proteolysis. J Biol Chem 291(37):19235–19244
Willem M et al (2015) Eta-secretase processing of APP inhibits neuronal activity in the hippocampus. Nature 526(7573):443–447
Tyan SH, Koo EH (2015) New tricks from an old dog: another synaptotoxic fragment from APP. Cell Res 25(11):1185–1186
Ahmad M et al (2006) Cleavage of amyloid-beta precursor protein (APP) by membrane-type matrix metalloproteinases. J Biochem 139(3):517–526
Baranger K et al (2016) MT5-MMP promotes Alzheimer’s pathogenesis in the frontal cortex of 5xFAD mice and APP trafficking in vitro. Front Mol Neurosci 9:163
Yong VW et al (2001) Metalloproteinases in biology and pathology of the nervous system. Nat Rev Neurosci 2(7):502–511
Visse R, Nagase H (2003) Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res 92(8):827–839
Wang X, Pei D (2001) Shedding of membrane type matrix metalloproteinase 5 by a furin-type convertase: a potential mechanism for down-regulation. J Biol Chem 276(38):35953–35960
Sekine-Aizawa Y et al (2001) Matrix metalloproteinase (MMP) system in brain: Identification and characterization of brain-specific MMP highly expressed in cerebellum. Eur J Neurosci 13(5):935–948
Tienari PJ et al (1996) Neuronal sorting and processing of amyloid precursor protein: implications for Alzheimer’s disease. Cold Spring Harb Symp Quant Biol 61:575–585
Zhang Z et al (2015) Delta-secretase cleaves amyloid precursor protein and regulates the pathogenesis in Alzheimer’s disease. Nat Commun 6:8762
Schonherr C et al (2016) Generation of aggregation prone N-terminally truncated amyloid beta peptides by meprin beta depends on the sequence specificity at the cleavage site. Mol Neurodegener 11:19
Jackle F et al (2015) Metalloprotease meprin beta is activated by transmembrane serine protease matriptase-2 at the cell surface thereby enhancing APP shedding. Biochem J 470(1):91–103
Arolas JL et al (2012) Structural basis for the sheddase function of human meprin beta metalloproteinase at the plasma membrane. Proc Natl Acad Sci U S A 109(40):16131–16136
Bien J et al (2012) The metalloprotease meprin beta generates amino terminal-truncated amyloid beta peptide species. J Biol Chem 287(40):33304–33313
Jefferson T et al (2011) Metalloprotease meprin beta generates nontoxic N-terminal amyloid precursor protein fragments in vivo. J Biol Chem 286(31):27741–27750
Sannerud R et al (2016) Restricted location of PSEN2/gamma-secretase determines substrate specificity and generates an intracellular Abeta pool. Cell 166(1):193–208
Meckler X, Checler F (2016) Presenilin 1 and presenilin 2 target gamma-secretase complexes to distinct cellular compartments. J Biol Chem 291(24):12821–12837
Kaether C et al (2006) Amyloid precursor protein and notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic 7(4):408–415
Skovronsky DM et al (2000) Protein kinase C-dependent alpha-secretase competes with beta-secretase for cleavage of amyloid-beta precursor protein in the trans-golgi network. J Biol Chem 275(4):2568–2575
Rocchi A et al (2003) Causative and susceptibility genes for Alzheimer’s disease: a review. Brain Res Bull 61(1):1–24
Haass C et al (2012) Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med 2(5):a006270
Caporaso GL et al (1994) Morphologic and biochemical analysis of the intracellular trafficking of the Alzheimer beta/A4 amyloid precursor protein. J Neurosci: Off J Soc Neurosci 14(5 Pt 2):3122–3138
Palacios G et al (1992) Beta-amyloid precursor protein localization in the Golgi apparatus in neurons and oligodendrocytes. An immunocytochemical structural and ultrastructural study in normal and axotomized neurons. Brain Res Mol Brain Res 15(3–4):195–206
Guo Q et al (2012) Amyloid precursor protein revisited: neuron-specific expression and highly stable nature of soluble derivatives. J Biol Chem 287(4):2437–2445
Das U et al (2013) Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron 79(3):447–460
Willnow TE, Andersen OM (2013) Sorting receptor SORLA—a trafficking path to avoid Alzheimer disease. J Cell Sci 126(Pt 13):2751–2760
Haass C et al (1992) Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature 357(6378):500–503
Cole GM et al (1992) An endosomal-lysosomal pathway for degradation of amyloid precursor protein. Ann N Y Acad Sci 674:103–117
Ehehalt R et al (2003) Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol 160(1):113–123
Schneider A et al (2008) Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J Neurosci 28(11):2874–2882
Lai A, Sisodia SS, Trowbridge IS (1995) Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain. J Biol Chem 270(8):3565–3573
Koo EH et al (1990) Precursor of amyloid protein in Alzheimer disease undergoes fast anterograde axonal transport. Proc Natl Acad Sci U S A 87(4):1561–1565
Amaratunga A, Fine RE (1995) Generation of amyloidogenic C-terminal fragments during rapid axonal transport in vivo of beta-amyloid precursor protein in the optic nerve. J Biol Chem 270(29):17268–17272
Morin PJ et al (1993) Amyloid precursor protein is synthesized by retinal ganglion cells, rapidly transported to the optic nerve plasma membrane and nerve terminals, and metabolized. J Neurochem 61(2):464–473
Kaether C, Skehel P, Dotti CG (2000) Axonal membrane proteins are transported in distinct carriers: a two-color video microscopy study in cultured hippocampal neurons. Mol Biol Cell 11(4):1213–1224
Yamazaki T, Selkoe DJ, Koo EH (1995) Trafficking of cell surface beta-amyloid precursor protein: retrograde and transcytotic transport in cultured neurons. J Cell Biol 129(2):431–442
Ferreira A, Caceres A, Kosik KS (1993) Intraneuronal compartments of the amyloid precursor protein. J Neurosci 13(7):3112–3123
Bloom GS et al (1988) Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry 27(9):3409–3416
DeBoer SR et al (2008) Conventional kinesin holoenzymes are composed of heavy and light chain homodimers. Biochemistry 47(15):4535–4543
Gyoeva FK, Bybikova EM, Minin AA (2000) An isoform of kinesin light chain specific for the Golgi complex. J Cell Sci 113(Pt 11):2047–2054
Cyr JL et al (1991) Molecular genetics of kinesin light chains: generation of isoforms by alternative splicing. Proc Natl Acad Sci U S A 88(22):10114–10118
Khodjakov A et al (1998) A specific light chain of kinesin associates with mitochondria in cultured cells. Mol Biol Cell 9(2):333–343
Morfini G et al (2016) Conventional kinesin: biochemical heterogeneity and functional implications in health and disease. Brain Res Bull 126(Pt 3):347–353
Lazarov O et al (2005) Axonal transport, amyloid precursor protein, kinesin-1, and the processing apparatus: revisited. J Neurosci 25(9):2386–2395
Matsuda S, Matsuda Y, D'Adamio L (2003) Amyloid beta protein precursor (AbetaPP), but not AbetaPP-like protein 2, is bridged to the kinesin light chain by the scaffold protein JNK-interacting protein 1. J Biol Chem 278(40):38601–38606
Inomata H et al (2003) A scaffold protein JIP-1b enhances amyloid precursor protein phosphorylation by JNK and its association with kinesin light chain 1. J Biol Chem 278(25):22946–22955
Chiba K et al (2014) Quantitative analysis of APP axonal transport in neurons: role of JIP1 in enhanced APP anterograde transport. Mol Biol Cell 25(22):3569–3580
Fu MM, Holzbaur EL (2013) JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J Cell Biol 202(3):495–508
Vagnoni A et al (2013) Loss of c-Jun N-terminal kinase-interacting protein-1 does not affect axonal transport of the amyloid precursor protein or abeta production. Hum Mol Genet 22(22):4646–4652
Rusu P et al (2007) Axonal accumulation of synaptic markers in APP transgenic drosophila depends on the NPTY motif and is paralleled by defects in synaptic plasticity. Eur J Neurosci 25(4):1079–1086
Dieckmann M, Dietrich MF, Herz J (2010) Lipoprotein receptors—an evolutionarily ancient multifunctional receptor family. Biol Chem 391(11):1341–1363
Nykjaer A, Willnow TE (2002) The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol 12(6):273–280
Pohlkamp T, Wasser CR, Herz J (2017) Functional roles of the interaction of APP and lipoprotein receptors. Front Mol Neurosci 10:54
Brown MS, Goldstein JL (1986) A receptor-mediated pathway for cholesterol homeostasis. Science 232(4746):34–47
May P, Bock HH, Herz J (2003) Integration of endocytosis and signal transduction by lipoprotein receptors. Sci STKE 2003(176):PE12
Wagner T, Pietrzik CU (2012) The role of lipoprotein receptors on the physiological function of APP. Exp Brain Res 217(3–4):377–387
Marcusson EG et al (1994) The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell 77(4):579–586
Jacobsen L et al (1996) Molecular characterization of a novel human hybrid-type receptor that binds the alpha2-macroglobulin receptor-associated protein. J Biol Chem 271(49):31379–31383
Yamazaki H et al (1996) Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J Biol Chem 271(40):24761–24768
Hermey G (2009) The Vps10p-domain receptor family. Cell Mol Life Sci: CMLS 66(16):2677–2689
Campbell ID, Spitzfaden C (1994) Building proteins with fibronectin type III modules. Structure 2(5):333–337
Quistgaard EM et al (2009) Ligands bind to sortilin in the tunnel of a ten-bladed beta-propeller domain. Nat Struct Mol Biol 16(1):96–98
Westergaard UB et al (2004) Functional organization of the sortilin Vps10p domain. J Biol Chem 279(48):50221–50229
Vogt L et al (2001) Calsyntenin-1, a proteolytically processed postsynaptic membrane protein with a cytoplasmic calcium-binding domain. Mol Cell Neurosci 17(1):151–166
Gul IS et al (2017) Evolution and diversity of cadherins and catenins. Exp Cell Res 358(1):3–9
Hintsch G et al (2002) The calsyntenins—a family of postsynaptic membrane proteins with distinct neuronal expression patterns. Mol Cell Neurosci 21(3):393–409
Pettem KL et al (2013) The specific alpha-neurexin interactor calsyntenin-3 promotes excitatory and inhibitory synapse development. Neuron 80(1):113–128
Araki Y et al (2007) The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J 26(6):1475–1486
Konecna A et al (2006) Calsyntenin-1 docks vesicular cargo to kinesin-1. Mol Biol Cell 17(8):3651–3663
Araki Y et al (2003) Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid beta-protein precursor metabolism. J Biol Chem 278(49):49448–49458
Herz J et al (1988) Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J 7(13):4119–4127
Willnow TE et al (1996) The low-density-lipoprotein receptor-related protein (LRP) is processed by furin in vivo and in vitro. Biochem J 313(Pt 1):71–76
Waldron E et al (2008) LRP1 modulates APP trafficking along early compartments of the secretory pathway. Neurobiol Dis 31(2):188–197
Jaeger S, Pietrzik CU (2008) Functional role of lipoprotein receptors in Alzheimer’s disease. Curr Alzheimer Res 5(1):15–25
Bu G (2009) Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci 10(5):333–344
Blacker D et al (1998) Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet 19(4):357–360
Roses AD (1996) Apolipoprotein E and Alzheimer’s disease. A rapidly expanding field with medical and epidemiological consequences. Ann N Y Acad Sci 802:50–57
Kang DE et al (2000) Modulation of amyloid beta-protein clearance and Alzheimer’s disease susceptibility by the LDL receptor-related protein pathway. J Clin Invest 106(9):1159–1166
Ulery PG et al (2000) Modulation of beta-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that LRP contributes to the pathogenesis of Alzheimer’s disease. J Biol Chem 275(10):7410–7415
Kounnas MZ et al (1995) LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell 82(2):331–340
Trommsdorff M et al (1998) Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem 273(50):33556–33560
Sandbrink R, Masters CL, Beyreuther K et al (1996) Ann N Y Acad Sci 777:281–287
Klug W et al (2011) Phosphorylation of LRP1 regulates the interaction with Fe65. FEBS Lett 585(20):3229–3235
Pietrzik CU et al (2004) FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J Neurosci: Off J Soc Neurosci 24(17):4259–4265
Li Y et al (2000) The YXXL motif, but not the two NPXY motifs, serves as the dominant endocytosis signal for low density lipoprotein receptor-related protein. J Biol Chem 275(22):17187–17194
Reekmans SM et al (2010) Inactivation of the proximal NPXY motif impairs early steps in LRP1 biosynthesis. Cell Mol Life Sci 67(1):135–145
Cam JA et al (2005) Rapid endocytosis of the low density lipoprotein receptor-related protein modulates cell surface distribution and processing of the beta-amyloid precursor protein. J Biol Chem 280(15):15464–15470
Pietrzik CU et al (2002) The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J 21(21):5691–5700
Koo EH, Squazzo SL (1994) Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem 269(26):17386–17389
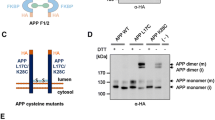
Herr UM et al (2017) LRP1 modulates APP intraneuronal transport and processing in its monomeric and dimeric state. Front Mol Neurosci 10:118
Eggert S et al (2017) Dimerization leads to changes in APP (amyloid precursor protein) trafficking mediated by LRP1 and SorLA. Cell Mol Life Sci. https://doi.org/10.1007/s00018-017-2625-7
Cam JA et al (2004) The low density lipoprotein receptor-related protein 1B retains beta-amyloid precursor protein at the cell surface and reduces amyloid-beta peptide production. J Biol Chem 279(28):29639–29646
Haas J et al (2011) LRP1b shows restricted expression in human tissues and binds to several extracellular ligands, including fibrinogen and apoE-carrying lipoproteins. Atherosclerosis 216(2):342–347
Strickland DK et al (1990) Sequence identity between the alpha 2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J Biol Chem 265(29):17401–17404
Kristensen T et al (1990) Evidence that the newly cloned low-density-lipoprotein receptor related protein (LRP) is the alpha 2-macroglobulin receptor. FEBS Lett 276(1–2):151–155
Liu CX et al (2001) The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J Biol Chem 276(31):28889–28896
Lane-Donovan C, Philips GT, Herz J (2014) More than cholesterol transporters: lipoprotein receptors in CNS function and neurodegeneration. Neuron 83(4):771–787
von Arnim CA et al (2005) The low density lipoprotein receptor-related protein (LRP) is a novel beta-secretase (BACE1) substrate. J Biol Chem 280(18):17777–17785
von Einem B et al (2010) The role of low-density receptor-related protein 1 (LRP1) as a competitive substrate of the amyloid precursor protein (APP) for BACE1. Exp Neurol 225(1):85–93
May P, Reddy YK, Herz J (2002) Proteolytic processing of low density lipoprotein receptor-related protein mediates regulated release of its intracellular domain. J Biol Chem 277(21):18736–18743
Lleo A et al (2005) Low density lipoprotein receptor-related protein (LRP) interacts with presenilin 1 and is a competitive substrate of the amyloid precursor protein (APP) for gamma-secretase. J Biol Chem 280(29):27303–27309
Deane R et al (2004) LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron 43(3):333–344
Deane R et al (2008) apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest 118(12):4002–4013
Rebeck GW et al (1993) Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron 11(4):575–580
Pflanzner T et al (2011) LRP1 mediates bidirectional transcytosis of amyloid-beta across the blood-brain barrier. Neurobiol Aging 32(12):2323 e1–2323 11
Storck SE et al (2016) Endothelial LRP1 transports amyloid-beta(1-42) across the blood-brain barrier. J Clin Invest 126(1):123–136
Shibata M et al (2000) Clearance of Alzheimer’s amyloid-ss(1-40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J Clin Invest 106(12):1489–1499
Sagare A et al (2007) Clearance of amyloid-beta by circulating lipoprotein receptors. Nat Med 13(9):1029–1031
Kanekiyo T, Bu G (2014) The low-density lipoprotein receptor-related protein 1 and amyloid-beta clearance in Alzheimer's disease. Front Aging Neurosci 6:93
Taira K et al (2001) LR11, a mosaic LDL receptor family member, mediates the uptake of ApoE-rich lipoproteins in vitro. Arterioscler Thromb Vasc Biol 21(9):1501–1506
Petersen CM et al (1997) Molecular identification of a novel candidate sorting receptor purified from human brain by receptor-associated protein affinity chromatography. J Biol Chem 272(6):3599–3605
Klinger SC et al (2011) SorLA regulates the activity of lipoprotein lipase by intracellular trafficking. J Cell Sci 124(Pt 7):1095–1105
Gliemann J et al (2004) The mosaic receptor sorLA/LR11 binds components of the plasminogen-activating system and platelet-derived growth factor-BB similarly to LRP1 (low-density lipoprotein receptor-related protein), but mediates slow internalization of bound ligand. Biochem J 381(Pt 1):203–212
Jacobsen L et al (2001) Activation and functional characterization of the mosaic receptor SorLA/LR11. J Biol Chem 276(25):22788–22796
Larsen JV et al (2016) Cytokine-like factor 1, an essential facilitator of cardiotrophin-like cytokine:ciliary neurotrophic factor receptor alpha signaling and sorLA-mediated turnover. Mol Cell Biol 36(8):1272–1286
Larsen JV, Petersen CM (2017) SorLA and CLC:CLF-1-dependent downregulation of CNTFRalpha as demonstrated by western blotting, inhibition of lysosomal enzymes, and immunocytochemistry. J Vis Exp. https://doi.org/10.3791/55019
Andersen OM et al (2006) Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry 45(8):2618–2628
Mehmedbasic A et al (2015) SorLA complement-type repeat domains protect the amyloid precursor protein against processing. J Biol Chem 290(6):3359–3376
Spoelgen R et al (2006) Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J Neurosci: Off J Soc Neurosci 26(2):418–428
Caglayan S et al (2014) Lysosomal sorting of amyloid-beta by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Science translational medicine 6(223):223ra20
Kitago Y et al (2015) Structural basis for amyloidogenic peptide recognition by sorLA. Nat Struct Mol Biol 22(3):199–206
Hermans-Borgmeyer I et al (1998) Unique expression pattern of a novel mosaic receptor in the developing cerebral cortex. Mech Dev 70(1–2):65–76
Andersen OM, Rudolph IM, Willnow TE (2016) Risk factor SORL1: from genetic association to functional validation in Alzheimer's disease. Acta Neuropathol 132(5):653–665
Scherzer CR et al (2004) Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol 61(8):1200–1205
Andersen OM et al (2005) Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A 102(38):13461–13466
Burgert T et al (2013) SORLA-dependent and -independent functions for PACS1 in control of amyloidogenic processes. Mol Cell Biol 33(21):4308–4320
Gustafsen C et al (2013) Sortilin and SorLA display distinct roles in processing and trafficking of amyloid precursor protein. J Neurosci: Off J Soc Neurosci 33(1):64–71
Klinger SC et al (2016) Polarized trafficking of the sorting receptor SorLA in neurons and MDCK cells. FEBS J 283(13):2476–2493
Hermey G et al (2006) Tumour necrosis factor alpha-converting enzyme mediates ectodomain shedding of Vps10p-domain receptor family members. Biochem J 395(2):285–293
Bohm C et al (2006) SorLA signaling by regulated intramembrane proteolysis. J Biol Chem 281(21):14547–14553
Nielsen MS et al (2007) Sorting by the cytoplasmic domain of the amyloid precursor protein binding receptor SorLA. Mol Cell Biol 27(19):6842–6851
Schmidt V et al (2007) SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem 282(45):32956–32964
Herskowitz JH et al (2012) GGA1-mediated endocytic traffic of LR11/SorLA alters APP intracellular distribution and amyloid-beta production. Mol Biol Cell 23(14):2645–2657
Jacobsen L et al (2002) The sorLA cytoplasmic domain interacts with GGA1 and -2 and defines minimum requirements for GGA binding. FEBS Lett 511(1–3):155–158
Dumanis SB et al (2015) Distinct functions for anterograde and retrograde sorting of SORLA in amyloidogenic processes in the brain. J Neurosci: Off J Soc Neurosci 35(37):12703–12713
Fjorback AW et al (2012) Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J Neurosci: Off J Soc Neurosci 32(4):1467–1480
Cullen PJ, Korswagen HC (2011) Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol 14(1):29–37
Muhammad A et al (2008) Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci U S A 105(20):7327–7332
Wen L et al (2011) VPS35 haploinsufficiency increases Alzheimer’s disease neuropathology. J Cell Biol 195(5):765–779
Holtzman DM, Herz J, Bu G (2012) Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med 2(3):a006312
Schmidt V et al (2012) Quantitative modelling of amyloidogenic processing and its influence by SORLA in Alzheimer’s disease. EMBO J 31(1):187–200
Huang TY et al (2016) SNX27 and SORLA interact to reduce amyloidogenic subcellular distribution and processing of amyloid precursor protein. J Neurosci: Off J Soc Neurosci 36(30):7996–8011
Clairfeuille T et al (2016) A molecular code for endosomal recycling of phosphorylated cargos by the SNX27-retromer complex. Nat Struct Mol Biol 23(10):921–932
Lauffer BE et al (2010) SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J Cell Biol 190(4):565–574
Grupe A et al (2006) A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet 78(1):78–88
Liang X et al (2009) Genomic convergence to identify candidate genes for Alzheimer disease on chromosome 10. Hum Mutat 30(3):463–471
Reitz C et al (2011) SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer’s disease risk. Ann Neurol 69(1):47–64
Wang HF et al (2012) SORCS1 and APOE polymorphisms interact to confer risk for late-onset Alzheimer’s disease in a northern Han Chinese population. Brain Res 1448:111–116
Xu W et al (2013) The genetic variation of SORCS1 is associated with late-onset Alzheimer’s disease in Chinese Han population. PLoS One 8(5):e63621
Hermey G et al (1999) Identification and characterization of SorCS, a third member of a novel receptor family. Biochem Biophys Res Commun 266(2):347–351
Savas JN et al (2015) The sorting receptor SorCS1 regulates trafficking of neurexin and AMPA receptors. Neuron 87(4):764–780
Traunmuller L et al (2016) Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science 352(6288):982–986
Lane RF et al (2010) Diabetes-associated SorCS1 regulates Alzheimer’s amyloid-beta metabolism: evidence for involvement of SorL1 and the retromer complex. J Neurosci: Off J Soc Neurosci 30(39):13110–13115
Hermey G et al (2015) SorCS1 variants and amyloid precursor protein (APP) are co-transported in neurons but only SorCS1c modulates anterograde APP transport. J Neurochem 135(1):60–75
Hermey G et al (2004) The three sorCS genes are differentially expressed and regulated by synaptic activity. J Neurochem 88(6):1470–1476
Hermey G, Schaller HC, Hermans-Borgmeyer I (2001) Transient expression of SorCS in developing telencephalic and mesencephalic structures of the mouse. Neuroreport 12(1):29–32
Oetjen S et al (2014) Spatiotemporal expression analysis of the growth factor receptor SorCS3. J Comp Neurol 522(15):3386–3402
Reitz C et al (2011) Impact of genetic variation in SORCS1 on memory retention. PLoS One 6(10):e24588
Hermey G et al (2003) Characterization of sorCS1, an alternatively spliced receptor with completely different cytoplasmic domains that mediate different trafficking in cells. J Biol Chem 278(9):7390–7396
Nielsen MS et al (2008) Different motifs regulate trafficking of SorCS1 isoforms. Traffic 9(6):980–994
Doray B et al (2012) Do GGA adaptors bind internal DXXLL motifs? Traffic 13(10):1315–1325
Clee SM et al (2006) Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat Genet 38(6):688–693
Goodarzi MO et al (2007) SORCS1: a novel human type 2 diabetes susceptibility gene suggested by the mouse. Diabetes 56(7):1922–1929
Granhall C et al (2006) High-resolution quantitative trait locus analysis reveals multiple diabetes susceptibility loci mapped to intervals <800 kb in the species-conserved Niddm1i of the GK rat. Genetics 174(3):1565–1572
Paterson AD et al (2010) A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes 59(2):539–549
Mazella J et al (1998) The 100-kDa neurotensin receptor is gp95/sortilin, a non-G-protein-coupled receptor. J Biol Chem 273(41):26273–26276
Sarret P et al (2003) Distribution of NTS3 receptor/sortilin mRNA and protein in the rat central nervous system. J Comp Neurol 461(4):483–505
Hermans-Borgmeyer I et al (1999) Expression of the 100-kDa neurotensin receptor sortilin during mouse embryonal development. Brain Res Mol Brain Res 65(2):216–219
Munck Petersen C et al (1999) Propeptide cleavage conditions sortilin/neurotensin receptor-3 for ligand binding. EMBO J 18(3):595–604
Westergaard UB et al (2005) SorCS3 does not require propeptide cleavage to bind nerve growth factor. FEBS Lett 579(5):1172–1176
Larsen JV et al (2014) Human sorCS1 binds sortilin and hampers its cellular functions. Biochem J 457(2):277–288
Nykjaer A et al (2004) Sortilin is essential for proNGF-induced neuronal cell death. Nature 427(6977):843–848
Teng HK et al (2005) ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J Neurosci: Off J Soc Neurosci 25(22):5455–5463
Larsen JV et al (2010) Sortilin facilitates signaling of ciliary neurotrophic factor and related helical type 1 cytokines targeting the gp130/leukemia inhibitory factor receptor beta heterodimer. Mol Cell Biol 30(17):4175–4187
Nielsen MS et al (1999) Sortilin/neurotensin receptor-3 binds and mediates degradation of lipoprotein lipase. J Biol Chem 274(13):8832–8836
Nilsson SK et al (2008) Endocytosis of apolipoprotein A-V by members of the low density lipoprotein receptor and the VPS10p domain receptor families. J Biol Chem 283(38):25920–25927
Carlo AS et al (2013) The pro-neurotrophin receptor sortilin is a major neuronal apolipoprotein E receptor for catabolism of amyloid-beta peptide in the brain. J Neurosci: Off J Soc Neurosci 33(1):358–370
Seaman MN (2007) Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J Cell Sci 120(Pt 14):2378–2389
Mari M et al (2008) SNX1 defines an early endosomal recycling exit for sortilin and mannose 6-phosphate receptors. Traffic 9(3):380–393
Canuel M et al (2008) Sortilin mediates the lysosomal targeting of cathepsins D and H. Biochem Biophys Res Commun 373(2):292–297
Corder EH et al (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261(5123):921–923
Yang M et al (2013) The intracellular domain of sortilin interacts with amyloid precursor protein and regulates its lysosomal and lipid raft trafficking. PLoS One 8(5):e63049
Soba P et al (2005) Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J 24(20):3624–3634
Koo EH et al (1996) Trafficking of cell-surface amyloid beta-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J Cell Sci 109(Pt 5):991–998
Burgos PV et al (2010) Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev Cell 18(3):425–436
Reitz C et al (2013) Independent and epistatic effects of variants in VPS10-d receptors on Alzheimer disease risk and processing of the amyloid precursor protein (APP). Transl Psychiatry 3:e256
Mufson EJ et al (2010) Preservation of cortical sortilin protein levels in MCI and Alzheimer’s disease. Neurosci Lett 471(3):129–133
Hu X et al (2017) Sortilin fragments deposit at senile plaques in human cerebrum. Front Neuroanat 11:45
Finan GM, Okada H, Kim TW (2011) BACE1 retrograde trafficking is uniquely regulated by the cytoplasmic domain of sortilin. J Biol Chem 286(14):12602–12616
Saadipour K et al (2013) Amyloid beta(1)(−)(4)(2) (Abeta(4)(2)) up-regulates the expression of sortilin via the p75(NTR)/RhoA signaling pathway. J Neurochem 127(2):152–162
Tan J, Evin G (2012) Beta-site APP-cleaving enzyme 1 trafficking and Alzheimer’s disease pathogenesis. J Neurochem 120(6):869–880
Nyborg AC et al (2006) Sortilin, SorCS1b, and SorLA Vps10p sorting receptors, are novel gamma-secretase substrates. Mol Neurodegener 1:3
Horiuchi K et al (2007) Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell 18(1):176–188
Evans SF et al (2011) Neuronal brain-derived neurotrophic factor is synthesized in excess, with levels regulated by sortilin-mediated trafficking and lysosomal degradation. J Biol Chem 286(34):29556–29567
Navarro V, Vincent JP, Mazella J (2002) Shedding of the luminal domain of the neurotensin receptor-3/sortilin in the HT29 cell line. Biochem Biophys Res Commun 298(5):760–764
Qi L et al (2011) Genetic risk score and risk of myocardial infarction in Hispanics. Circulation 123(4):374–380
Takeuchi F et al (2012) Association of genetic variants influencing lipid levels with coronary artery disease in Japanese individuals. PLoS One 7(9):e46385
Arvind P et al (2014) CELSR2-PSRC1-SORT1 gene expression and association with coronary artery disease and plasma lipid levels in an Asian Indian cohort. J Cardiol 64(5):339–346
Jones GT et al (2013) A sequence variant associated with sortilin-1 (SORT1) on 1p13.3 is independently associated with abdominal aortic aneurysm. Hum Mol Genet 22(14):2941–2947
Lee JY et al (2013) A genome-wide association study of a coronary artery disease risk variant. J Hum Genet 58(3):120–126
Angelakopoulou A et al (2012) Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: cardiovascular biomarker genetics collaboration. Eur Heart J 33(3):393–407
Stampfer MJ (2006) Cardiovascular disease and Alzheimer’s disease: common links. J Intern Med 260(3):211–223
Andersson CH et al (2016) A genetic variant of the sortilin 1 gene is associated with reduced risk of Alzheimer’s disease. J Alzheimers Dis: JAD 53(4):1353–1363
Zeng F et al (2013) No association of SORT1 gene polymorphism with sporadic Alzheimer’s disease in the Chinese Han population. Neuroreport 24(9):464–468
Lambert JC et al (2013) Genome-wide haplotype association study identifies the FRMD4A gene as a risk locus for Alzheimer’s disease. Mol Psychiatry 18(4):461–470
Naj AC et al (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43(5):436–441
Beecham GW et al (2014) Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet 10(9):e1004606
Reynolds CA et al (2010) Analysis of lipid pathway genes indicates association of sequence variation near SREBF1/TOM1L2/ATPAF2 with dementia risk. Hum Mol Genet 19(10):2068–2078
Rogaeva E et al (2007) The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet 39(2):168–177
Ward ME, Miller BL (2011) Potential mechanisms of progranulin-deficient FTLD. J Mol Neurosci: MN 45(3):574–582
Rademakers R, Neumann M, Mackenzie IR (2012) Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol 8(8):423–434
Bird T et al (2003) Epidemiology and genetics of frontotemporal dementia/Pick’s disease. Ann Neurol 54(Suppl 5):S29–S31
Hu F et al (2010) Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 68(4):654–667
Carrasquillo MM et al (2010) Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet 87(6):890–897
Zheng Y et al (2011) C-terminus of progranulin interacts with the beta-propeller region of sortilin to regulate progranulin trafficking. PLoS One 6(6):e21023
Gendron TF, Rademakers R, Petrucelli L (2013) TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43. J Alzheimers Dis 33(Suppl 1):S35–S45
Polymenidou M et al (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14(4):459–468
Prudencio M et al (2012) Misregulation of human sortilin splicing leads to the generation of a nonfunctional progranulin receptor. Proc Natl Acad Sci U S A 109(52):21510–21515
Rogelj B et al (2006) The X11/mint family of adaptor proteins. Brain Res Rev 52(2):305–315
Um JW et al (2014) Calsyntenins function as synaptogenic adhesion molecules in concert with neurexins. Cell Rep 6(6):1096–1109
Araki Y et al (2004) Coordinated metabolism of Alcadein and amyloid beta-protein precursor regulates FE65-dependent gene transactivation. J Biol Chem 279(23):24343–24354
Hata S et al (2009) Alcadein cleavages by amyloid beta-precursor protein (APP) alpha- and gamma-secretases generate small peptides, p3-Alcs, indicating Alzheimer disease-related gamma-secretase dysfunction. J Biol Chem 284(52):36024–36033
Maruta C et al (2012) Constitutive cleavage of the single-pass transmembrane protein alcadeinalpha prevents aberrant peripheral retention of Kinesin-1. PLoS One 7(8):e43058
Hata S et al (2011) Alternative processing of gamma-secretase substrates in common forms of mild cognitive impairment and Alzheimer’s disease: evidence for gamma-secretase dysfunction. Ann Neurol 69(6):1026–1031
Vagnoni A et al (2012) Calsyntenin-1 mediates axonal transport of the amyloid precursor protein and regulates Abeta production. Hum Mol Genet 21(13):2845–2854
Ringman JM et al (2012) Proteomic changes in cerebrospinal fluid of presymptomatic and affected persons carrying familial Alzheimer disease mutations. Arch Neurol 69(1):96–104
Uchida Y et al (2013) Calsyntenin-3 C-terminal fragment accumulates in dystrophic neurites surrounding abeta plaques in tg2576 mouse and Alzheimer disease brains: Its neurotoxic role in mediating dystrophic neurite formation. Am J Pathol 182(5):1718–1726
Ludwig A et al (2009) Calsyntenins mediate TGN exit of APP in a kinesin-1-dependent manner. Traffic 10(5):572–589
Vagnoni A et al (2011) Phosphorylation of kinesin light chain 1 at serine 460 modulates binding and trafficking of calsyntenin-1. J Cell Sci 124(Pt 7):1032–1042
Steuble M et al (2010) Molecular characterization of a trafficking organelle: dissecting the axonal paths of calsyntenin-1 transport vesicles. Proteomics 10(21):3775–3788
Goldsbury C et al (2006) Inhibition of APP trafficking by tau protein does not increase the generation of amyloid-beta peptides. Traffic 7(7):873–888
Stamer K et al (2002) Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J Cell Biol 156(6):1051–1063
Borg JP et al (1998) The X11alpha protein slows cellular amyloid precursor protein processing and reduces Abeta40 and Abeta42 secretion. J Biol Chem 273(24):14761–14766
Tomita S et al (1999) Interaction of a neuron-specific protein containing PDZ domains with Alzheimer’s amyloid precursor protein. J Biol Chem 274(4):2243–2254
Shrivastava-Ranjan P et al (2008) Mint3/X11gamma is an ADP-ribosylation factor-dependent adaptor that regulates the traffic of the Alzheimer’s precursor protein from the trans-Golgi network. Mol Biol Cell 19(1):51–64
Brunholz S et al (2011) Axonal transport of APP and the spatial regulation of APP cleavage and function in neuronal cells. Exp Brain Res. 217(3–4):353–364. https://doi.org/10.1007/s00221-011-2870-1
Grbovic OM et al (2003) Rab5-stimulated up-regulation of the endocytic pathway increases intracellular beta-cleaved amyloid precursor protein carboxyl-terminal fragment levels and Abeta production. J Biol Chem 278(33):31261–31268
Ikin AF et al (1996) Alzheimer amyloid protein precursor is localized in nerve terminal preparations to Rab5-containing vesicular organelles distinct from those implicated in the synaptic vesicle pathway. J Biol Chem 271(50):31783–31786
Siddiqui TJ, Craig AM (2011) Synaptic organizing complexes. Curr Opin Neurobiol 21(1):132–143
Schilling S et al (2017) APLP1 is a synaptic cell adhesion molecule, supporting maintenance of dendritic spines and basal synaptic transmission. J Neurosci 37:5345–5365
Stahl R et al (2014) Shedding of APP limits its synaptogenic activity and cell adhesion properties. Front Cell Neurosci 8:410
Baumkotter F et al (2014) Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain. J Neurosci: Off J Soc Neurosci 34(33):11159–11172
Wang Z et al (2009) Presynaptic and postsynaptic interaction of the amyloid precursor protein promotes peripheral and central synaptogenesis. J Neurosci: Off J Soc Neurosci 29(35):10788–10801
Dean C, Dresbach T (2006) Neuroligins and neurexins: linking cell adhesion, synapse formation and cognitive function. Trends Neurosci 29(1):21–29
Dalva MB, McClelland AC, Kayser MS (2007) Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci 8(3):206–220
Lu Z et al (2014) Calsyntenin-3 molecular architecture and interaction with neurexin 1alpha. J Biol Chem 289(50):34530–34542
Scheff SW, Price DA (2006) Alzheimer’s disease-related alterations in synaptic density: neocortex and hippocampus. J Alzheimers Dis: JAD 9(3 Suppl):101–115
Mhatre SD et al (2015) Microglial malfunction: the third rail in the development of Alzheimer’s disease. Trends Neurosci 38(10):621–636
Thomas DM, Francescutti-Verbeem DM, Kuhn DM (2006) Gene expression profile of activated microglia under conditions associated with dopamine neuronal damage. FASEB J 20(3):515–517
Jehle AW et al (2006) ATP-binding cassette transporter A7 enhances phagocytosis of apoptotic cells and associated ERK signaling in macrophages. J Cell Biol 174(4):547–556
Villegas-Llerena C et al (2016) Microglial genes regulating neuroinflammation in the progression of Alzheimer’s disease. Curr Opin Neurobiol 36:74–81
Southam KA, Vincent AJ, Small DH (2016) Do microglia default on network maintenance in Alzheimer’s disease? J Alzheimers Dis 51(3):657–669
Acknowledgments
We thank Prof. Claus Pietrzik for critically reading the manuscript.
Funding
Funding for SE was from the TU Nachwuchsring (University of Kaiserslautern) and SK and GH were supported by Alzheimer Forschung Initiative (AFI).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Eggert, S., Thomas, C., Kins, S. et al. Trafficking in Alzheimer’s Disease: Modulation of APP Transport and Processing by the Transmembrane Proteins LRP1, SorLA, SorCS1c, Sortilin, and Calsyntenin. Mol Neurobiol 55, 5809–5829 (2018). https://doi.org/10.1007/s12035-017-0806-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-017-0806-x