Abstract
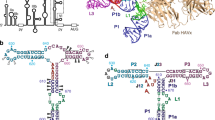
The self-cleaving ribozyme of the hepatitis delta virus (HDV) is the only catalytic RNA known to be required for the viability of a human pathogen. We obtained crystals of a 72-nucleotide, self-cleaved form of the genomic HDV ribozyme that diffract X-rays to 2.3 Å resolution by engineering the RNA to bind a small, basic protein without affecting ribozyme activity. The co-crystal structure shows that the compact catalytic core comprises five helical segments connected as an intricate nested double pseudoknot. The 5′-hydroxyl leaving group resulting from the self-scission reaction is buried deep within an active-site cleft produced by juxtaposition of the helices and five strand-crossovers, and is surrounded by biochemically important backbone and base functional groups in a manner reminiscent of protein enzymes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lai, M. M. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 64, 259–286 (1995).
Been, M. D. & Wickham, G. S. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur. J. Biochem. 247, 741–753 (1997).
Kuo, M. Y.-P., Sharmeen, L., Dinter-Gottlieb, G. & Taylor, J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol. 62, 4439–444 (1988).
Sharmeen, L., Kuo, M. Y.-P., Dinter-Gottlieb, G. & Taylor, J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 62, 2674–2679 (1988).
Wu, H.-N. et al. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc. Natl Acad. Sci. USA 86, 1831–1835 (1989).
Perrotta, A. T. & Been, M. D. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 18, 6821–6827 (1990).
Perrotta, A. T. & Been, M. D. Apseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature 350, 434–436 (1991).
Tanner, N. K. et al. Athree-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr. Biol. 4, 488–498 (1994).
Thill, G., Vasseur, M. & Tanner, N. K. Structural and sequence elements required for the self-cleaving activity of the hepatitis delta virus ribozyme. Biochemistry 32, 4254–4262 (1993).
Pley, H. W., Flaherty, K. M. & McKay, D. B. Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74 (1994).
Scott, W. G., Finch, J. T. & Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell 81, 991–1002 (1995).
Richards, F. M. & Wyckoff, H. W. in The Enzymes (ed. Boyer, P. D.) 647–806 (Academic, New York, 1971).
Rosenstein, S. P. & Been, M. D. Self-cleavage of hepatitis delta virus genomic strand RNA is enhanced under partially denaturing conditions. Biochemistry 29, 8011–8016 (1990).
Duhamel, J. et al. Secondary structure content of the HDV ribozyme in 95% formamide. Nucleic Acids Res. 24, 3911–3917 (1996).
Suh, Y.-A., Kumar, P. K. R., Taira, K. & Nishikawa, S. Self-cleavage activity of the genomic HDV ribozyme in the presence of various divalent metal ions. Nucleic Acids Res. 21, 3277–3280 (1993).
Ferré-D'Amaré, A. R., Zhou, K. & Doudna, J. A. Ageneral module for RNA crystallization. J. Mol. Biol. 279, 621–631 (1998).
Oubridge, C., Ito, N. Evans, P. R., Teo, C.-H. & Nagai, K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372, 432–438 (1994).
Kolk, M. H. et al. NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science 280, 434–438 (1998).
Tang, C. K. & Draper, D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell 57, 531–536 (1989).
Gluick, T. C. & Draper, D. E. Thermodynamics of folding a pseudoknotted mRNA fragment. J. Mol. Biol. 241, 246–262 (1994).
Ekland, E. H., Szostak, J. W. & Bartel, D. P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269, 364–370 (1995).
Kolk, M. H., Heus, H. A. & Hilbers, C. W. The structure of the isolated, central hairpin of the HDV antigenomic ribozyme: novel structural features and similarity of the loop in the ribozyme and free in solution. EMBO J. 16, 3685–3692 (1997).
Lynch, S. R. & Tinoco, I. J The structure of the L3 loop from the hepatitis delta virus ribozyme: a syn cytidine. Nucleic Acids Res. 26, 980–987 (1998).
Perrotta, A. T. & Been, M. D. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 24, 1314–1321 (1996).
Rosenstein, S. P. & Been, M. D. Hepatitis delta virus ribozymes fold to generate a solvent-inaccessible core with essential nucleotides near the cleavage site phosphate. Biochemistry 35, 11403–11413 (1996).
Nishikawa, F., Fauzi, H. & Nishikawa, S. Detailed analysis of base preferences at the cleavage site of a trans-acting HDV ribozyme: a mutation that changes cleavage site specificity. Nucleic Acids Res. 25, 1605–1610 (1997).
Strobel, S. A. & Cech, T. R. Minor groove recognition of the conserved G-U pair at the Tetrahymena ribozyme reaction site. Science 267, 675–679 (1995).
Cate, J. H. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 (1996).
Pley, H. W., Flaherty, K. M. & McKay, D. B. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature 372, 111–113 (1994).
Kumar, P. K. R. et al. Random mutations to evaluate the role of bases at two important single-stranded regions of the genomic HDV ribozyme. Nucleic Acids Res. 20, 3919–3924 (1992).
Kumar, P. K. R., Suh, Y.-A., Taira, K. & Nishikawa, S. Point and compensation mutations to evaluate essential stem structures of genomic HDV ribozyme. FASEB J. 7, 124–129 (1993).
Kumar, P. K. R., Taira, K. & Nishikawa, S. Chemical probing studies of variants of the genomic hepatitis delta virus ribozyme by primer extension analysis. Biochemistry 33, 583–592 (1994).
Belinsky, M. G., Britton, E. & Dinter-Gottlieb, G. Modification interference analysis of a self-cleaving RNA from hepatitis delta virus. FASEB J. 7, 130–136 (1993).
Wadkins, T. S. & Been, M. D. Core-associated non-duplex sequences distinguishing the genomic and antigenomic self-cleaving RNAs of hepatitis delta virus. Nucleic Acids Res. 25, 4085–4092 (1997).
Rosenstein, S. P. & Been, M. D. Evidence that genomic and antigenomic RNA self-cleaving elements from hepatitis delta virus have similar secondary structures. Nucleic Acids Res. 19, 5409–5416 (1991).
Huang, Z.-S., Ping, Y.-H. & Wu, H.-N. An AU at the first base pair of helix 3 elevates the catalytic activity of hepatitis delta virus ribozymes. FEBS lett. 413, 299–303 (1997).
Jeoung, Y.-H., Kumar, P. K. R., Suh, Y.-A., Taira, K. & Nishikawa, S. Identification of phosphate oxygens that are important for self-cleavage activity of the HDV ribozyme by phosphorothioate substitution interference analysis. Nucleic Acids Res. 22, 3722–3727 (1994).
Bravo, C., Lescure, F., Laugâa, P., Fourrey, J.-L. & Favre, A. Folding of the HDV antigenomic ribozyme pseudoknot structure deduced from long-range photocrosslinks. Nucleic Acids Res. 24, 1351–1359 (1996).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–776 (1994).
Terwilliger, T. C. & Kim, S.-H. Generalized method of determining heavy-atom positions using the difference Patterson function. Acta Crystallogr. A 43, 1–5 (1987).
de la Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997).
Abrahams, J. P. & Leslie, A. G. W. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A. T. et al. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Brünger, A. T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 (1992).
Laskowski, R. J., Macarthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–290 (1993).
Pleij, C. W. A., Rietveld, K. & Bosch, L. Anew principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 13, 1717–1731 (1985).
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insight from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991).
Carson, M. Ribbons 2.0. J. Appl. Crystallogr. 24, 958–961 (1991).
Acknowledgements
We thank K. Nagai for U1A-RBD expression plasmids; C. Ogata and D. Cook for help at beamline X4A of the National Synchrotron Light Source (Brookhaven National Laboratory); the staff of the Cornell High Energy Synchrotron Source for support at beamlines A-1 and F-1; R. Batey, D.Battle, J. Kieft, A. Luptak and R. Rambo for help with synchrotron data collection and useful comments; P. Adams, M. Been, S. Bellon, C. Correll, R. Gaudet, D. Herschlag, J. Ippolito, P. Moore, L.Silvian, T. Steitz, S. Strobel, O. Uhlenbeck and D. Wilson for helpful discussions; and the staff of the Yale Center for Structural Biology for crystallographic and computational support. A.R.F. was a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. This work was supported in part by grants from the Jane Coffin Childs Memorial Fund for Medical Research, the Searle Scholars program, the NIH, and a Beckman Young Investigator award. J.A.D. is an assistant investigator of the Howard Hughes Medical Institute, a Searle scholar, and a Fellow of the David and Lucile Packard Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ferré-D'Amaré, A., Zhou, K. & Doudna, J. Crystal structure of a hepatitis delta virus ribozyme. Nature 395, 567–574 (1998). https://doi.org/10.1038/26912
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/26912
This article is cited by
-
Targeted mRNA demethylation in Arabidopsis using plant m6A editor
Plant Methods (2023)
-
Ribozyme-mediated CRISPR/Cas9 gene editing in pyrethrum (Tanacetum cinerariifolium) hairy roots using a RNA polymerase II-dependent promoter
Plant Methods (2022)
-
Structure of active human telomerase with telomere shelterin protein TPP1
Nature (2022)
-
Hepatitis D: challenges in the estimation of true prevalence and laboratory diagnosis
Gut Pathogens (2021)
-
Cellular conditions of weakly chelated magnesium ions strongly promote RNA stability and catalysis
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.