Abstract
EZH2 is the catalytic subunit of the polycomb repressive complex 2 (PRC2), which is a highly conserved histone methyltransferase that methylates lysine 27 of histone 3. Overexpression of EZH2 has been found in a wide range of cancers, including those of the prostate and breast. In this review, we address the current understanding of the oncogenic role of EZH2, including its PRC2-dependent transcriptional repression and PRC2-independent gene activation. We also discuss the connections between EZH2 and other silencing enzymes, such as DNA methyltransferase and histone deacetylase. We comprehensively address the architecture of the PRC2 complex and the crucial roles of each subunit. Finally, we summarize new progress in developing EZH2 inhibitors, which could be a new epigenetic therapy for cancers.
Similar content being viewed by others
Introduction
In eukaryotes, the posttranslational modifications of histones play crucial roles in regulating chromatin structure and gene expression. This epigenetic alteration is different from the usual genetic alterations because once the DNA sequence is altered by mutations, it is very difficult to restore the former sequence. However, epigenetic changes can be reversed by specific inhibitors that target enzymes such as DNA methyltransferase, histone methyltransferase, and histone deacetylase1. Because epigenetic abnormalities are common in human cancer and play a key role in tumor progression, an understanding of epigenetic alterations can benefit the process of drug design and discovery for cancer treatment2.
EZH2, as a catalytic subunit of the polycomb repressive complex 2 (PRC2), represses gene expression by methylating lysine 27 of histone 3 (H3K27). EZH2-mediated methylation is a potential independent mechanism for epigenetic silencing of tumor suppressor genes in cancer. Recent studies have found that EZH2 is overexpressed in a wide range of cancers such as those of the prostate, breast, and bladder, which makes EZH2 an attractive anti-cancer drug target. In this review, we focus on the PRC2 machinery, including new advances in understanding how the five PRC2 subunits interact with each other. We also review new findings that suggest that EZH2 acts as transcriptional gene activator rather than a gene silencer. New progress on inhibitor design has been made by targeting the conserved SET domain of EZH2, which may contribute to the development of novel treatment strategies against different cancers.
Biological function of EZH2
PRC2 is an essential chromatin modifier that is conserved across organisms from plants to flies and humans3. This complex represses the transcription of target genes through trimethylation of lysine 27 on histone 3 (H3K27me3), which is currently viewed as its predominant function in vivo4. The human PRC2 complex includes five subunits: EZH2, EED, SUZ12, RbAp46/48, and AEBP2 (Figure 1). A common biological function of PRC2 is the transcriptional silencing of genes involved in differentiation.
Possible architecture of the PRC2 complex. (A) Models of the fly and human PRC2 complexes. The subunits and interactions between them are shown. (B) Domain organizations of each subunit in the human PRC2 complex. Domain “1”, binding region for PHF1 in human cells and PCL in flies; domain “2”, binding region for SUZ12; CXC, cysteine-rich domain; SANT, domain that allows chromatin remodeling protein to interact with histones; SET, catalytic domain of EZH2; VEFS, VRN2-EMF2-FIS2-SUZ12 domain; WD, WD-40 domain; WDB, WD-40 binding domain; Zn, Zn-finger region.
PRC2 catalyzes three sequential methylation reactions at H3K27, resulting in mono-, di-, and trimethylated H3K27 (H3K27me1, H3K27me2, and H3K27me3). EZH2 is the catalytic subunit of the PRC2 complex, and its C-terminal SET domain exhibits methyltransferase activity. EZH2 lacks enzymatic function on its own; however, it gains robust histone lysine methyltransferase activity when it complexes with two other noncatalytic subunits of the PRC2 complex, namely the zinc-finger-containing SUZ12 and the WD40-repeat protein EED5. These two subunits are required to maintain the integrity of PRC2, and mutations in either gene may destabilize EZH26,7,8.
Regarding the other subunits, it has been reported that the PRC2 subcomplex without the RbAp48 subunit can maintain substantial enzymatic activity, suggesting that the RbAp48 subunit is not required for the histone methyltransferase activity of EZH23. The AEBP2 subunit acts as a cofactor that interacts with the other four subunits by binding to the center of the PRC2 complex, which helps to stabilize the overall architecture of PRC2. The AEBP2 subunit also facilitates the PRC2 complex targeting to specific DNA sites and enhances its methyltransferase activity.
EZH2 as an epigenetic silencer
Currently, EZH2 is believed to function predominately as a transcriptional repressor that silences an array of target genes, including more than 200 tumor suppressors5. This view is supported by the finding that many PRC2-repressed genes are linked to poor outcomes in prostate cancer patients9. It is known that the PRC1 complex, which comprises B lymphoma Mo-MLV insertion region 1 and the ring finger proteins RING1 and RING2, functions cooperatively with PRC2 during epigenetic silencing by ubiquitinating lysine 119 of histone H2A10. The PRC2-mediated trimethylation of H3K27 is believed to recruit PRC1 to target gene loci, leading to a more condensed chromatin configuration. In support of this coordinated recruitment and function, the expression of PRC1 and PRC2 proteins can be integrated through a network of regulatory microRNAs11, such as miR-200, which can be transcriptionally repressed by EZH2. Because these miRNAs are involved in the regulation of PRC1, their repression by EZH2 can upregulate PRC1. In summary, it is suggested that there is a molecular link between these two protein complexes, which play important roles in regulating the state of chromatin and therefore affect the transcription of target genes, through the miRNA network.
Cooperation with other epigenetic silencing enzymes
The EZH2 histone methyltransferase usually cooperates with other epigenetic silencing enzymes. Recent studies have shown that there are physical and functional links between EZH2, DNA methyltransferases (DNMTs)12, and histone deacetylases (HDACs)13,14, suggesting the potential interplay between these different classes of epigenetic modulation enzymes in the control of gene expression.
Figure 2 illustrates a model for the cooperation of epigenetic silencing enzymes. Initially, the PRC2 complex methylates lysine 27 on histone 3 (H3K27me) and silences target genes. However, if K27 is acetylated, a histone deacetylase may be required. HDACs can help to modify the local histone code for silencing by deacetylating H3K27 and other lysines including H3K29, H3K14, and H4K8, thereby making the ε-amino group of lysine side chains available for methylation by PRC2. PRC2 can also cooperate with DNMTs to convert the target genes into a more deeply and permanently silenced chromatin state. During cellular transformation, certain genes that acquire methylation marks due to the action of the PRC2 complex will later become CpG-hypermethylated. Thus, the PRC2 complex can cooperate with HDACs to alter histone marks from acetylation to methylation, and it can also recruit DNMTs to produce a denser chromatin state. Functional links between EZH2, HDACs, and DNMTs have been found in colon, prostate, liver, lung, ovarian, and breast tumors, and all three types of epigenetic silencing machinery may contribute to the control of abnormal gene expression in cancer cells5.
A model for the collaboration of epigenetic silencing enzymes, including core of the PRC2 complex, DNA methyltransferase (DNMT), and histone deacetylase (HDAC). In this model, if K27 is pre-acetylated, HDAC may first deacetylates it, and then the target genes are silenced through the methylation of K27 by PRC2. DNMTs may also be recruited by PRC2, and after methylating CpG DNA of target genes, making the chromatin state more deeply silenced. Ac, acetylation; and Me, methylation.
Mutations in EZH2
EZH2 has been reported to harbor various heterozygous mutations at tyrosine 641 (Y641) in the C-terminal SET domain; such mutations are found in 7% of follicular lymphomas and 22% of germinal center B-cell and diffuse large B-cell lymphomas (DLBCLs)15. Although initially reported to be a loss-of-function mutation, subsequent studies have shown that the Y641 mutation actually results in a gain of function16,17. In contrast to the wild type, which has a substrate preference for unmethylated H3K27 and monomethylated H3K27me1, the Y641 mutants (including Y641F, Y641N, Y641S, Y641H, and Y641C) have enhanced catalytic efficiency for dimethylated H3K27me216,17. Thus, both the wild type and the Y641 mutants can work together to increase the levels of H3K27me3.
Because the crystal structure of EZH2 is not yet available, a homology model was constructed to predict the substrate specificities of the wild type and the Y641 mutant18. The model shows that there is little room in the wild-type binding pocket for dimethylated lysine to rotate into the position to accept a third methyl group. Thus, the Y641 residue may play two roles: first, it may participate in the orientation of unmethylated and monomethylated lysine, and second, it may sterically restrict activity with a dimethylated substrate.
Another mutant, A677G, is also found in lymphoma cell lines, albeit at a low frequency (below 2%–3%). Characterization of this mutant protein has shown that the replacement of alanine 677 with glycine leads to increased activity with H3K27me2 substrates. This result is similar to that obtained with the Y641 mutant; however, mechanistic differences are evident. Relative to the Y641 mutant, which loses its activity with nonmethylated H3K27 substrate, the A677G mutant retains the crucial interactions with H3K27 substrate that are present in the wild-type EZH2, leading to efficient use of all three methylation substrates (H3K27, H3K27me1, and H3K27me2).
EZH2 functions independently as a gene activator
In addition to its role as a transcriptional repressor, several studies have shown that EZH2 may also function in target gene activation19,20,21. Recently, Xu et al reported that EZH2 plays an important role in castration-resistant prostate cancer, and its oncogenic function does not depend on silencing but rather on transcriptional induction of its target genes21. These authors found that a subset of EZH2-bound genes did not bind the PRC2 subunit SUZ12 or display H3K27me3. Many of these genes were downregulated upon EZH2 knockdown, suggesting that the role of EZH2 as an activator was independent of the PRC2 complex. Xu et al also showed that the methyltransferase activity of EZH2 was required for both EZH2-dependent gene activation and androgen-independent growth, which differs from the findings of early reports indicating that EZH2 functions as a gene activator19,20. The latter findings were observed in breast cancer cells, where EZH2 activates NF-κB target genes through the formation of a ternary complex with the NF-κB components RelA and RelB that does not require other PRC2 subunits19. EZH2 overexpression can also lead to its interaction with Wnt signaling components and subsequent activation of the c-myc and cyclin D1 genes; again, this function is independent of its methyltransferase activity20. It has been suggested that EZH2 may act as a multifaceted molecule; ie, it may function as either a transcriptional repressor or activator in promoting breast tumorigenesis22.
In castration-resistant prostate cancer, EZH2-mediated transcriptional activation may occur via methylation of the androgen receptor (AR) or other associated proteins, which would indicate a new role for EZH2 in the methylation of nonhistone proteins. Xu et al21 showed that phosphorylation of EZH2 at Ser21, mediated directly or indirectly by the PI3K-Akt pathway, can alter its function from a polycomb repressor to a transcriptional co-activator of AR and (potentially) other proteins (Figure 3). This finding indicates the potential for the development of inhibitors that can specifically target the activator function of EZH2 while sparing its PRC2 repressive function.
A model of the EZH2 functional switch from a polycomb repressor to a transcriptional activator in castration-resistant prostate cancer. PI3K/AKT pathway activation can lead to phosphorylation of EZH2 at Ser21. This phosphorylation event shifts EZH2 from a transcriptional repressor associated with PRC2 to a transcriptional co-activator cooperating with AR, using an intact SET methyltransferase domain.
EZH2 and cancer
Hyperactivation of EZH2, either by overexpression or mutations, is found in a variety of malignancies including prostate, breast, uterine, gastric, and renal cell cancers in addition to melanoma5. EZH2 expression is correlated with aggressiveness, metastasis, and poor prognosis in most of these cancers. A recent study also found that EZH2 was overexpressed in non-small cell lung cancers and lymphoma23. The functional consequence of increased EZH2 in cancer tissues includes the silencing of genes that promote differentiation and restrain proliferation. Table 1 summarizes the extensive reports describing EZH2 hyperactivation in various cancers.
EZH2 in prostate cancer
A gene profiling study showed that EZH2 upregulated oncogenes in metastatic prostate cancer and that loss of EZH2 inhibited the growth of prostate cancer cells25. According to this study, EZH2 overexpression could be a valuable prognostic indicator of patient outcome. Another recent report focused on the relationship between EZH2 and androgen signaling pathways24, showing that EZH2 expression is repressed by androgens. This repression requires a functional androgen receptor and is mediated through retinoblastoma (RB)/p130-dependent pathways. Further, EZH2 may activate AR-repressed genes in an androgen-deprived environment because EZH2 is frequently overexpressed in metastatic prostate cancers35.
EZH2 in breast cancer
Breast cancer is the most common malignancy and the second leading cause of cancer-related death among women. Abnormally elevated EZH2 levels have been found to be highly correlated with invasiveness and increased proliferation rates of breast carcinomas26,30. These studies also proposed that EZH2 is a promising biomarker for aggressive breast cancers with poor prognosis and that it can be an independent indicator of clinical outcome. Overexpression of EZH2 is correlated with many signaling pathways in breast cancer, such as the pRB-E2F, PI3K/Akt, and estrogen receptor pathways27,28,29. For example, Gonzales et al showed that EZH2 overexpression in breast cancer cells can activate the PI3K/Akt pathway, especially through activation of the Akt isoform28. Based on experimental evidence, Deb et al proposed that EZH2 may function as a co-activator when it is overexpressed during malignancy and that it can be recruited to the estrogen signaling pathway to enhance estrogen signaling and promote proliferation22.
EZH2 in B-cell lymphomas
Lymphogenesis represents a special case wherein EZH2 is repressed in resting naive B cells but is highly upregulated in primary lymphoid follicles during B cell activation and germinal center (GC) formation23. EZH2 is overexpressed in GC-derived lymphomas, such as DLBCL32. Moreover, mutations in the SET domain of EZH2 that favor the formation of trimethylated H3K27 — such as Y641F — have been frequently identified in both DLBCL and follicular lymphoma15,31. In addition, DLBCLs are dependent on the oncogenic function of EZH2 independent of its mutational state because impairments in PRC2 enzyme activity can abolish tumorigenesis by both mutant and wild-type cancer cells. Thus, EZH2 is a promising drug target that can be specifically inhibited by small molecules (see below).
The PRC2 complex and related structures
Molecular architecture of the PRC2 complex
Figure 1 illustrates the domain organization of PRC2 and the composition of individual subunits. The multiple partners of the PRC2 complex indicate its highly cooperative nature, which is essential for its function. Comprehensive studies have been performed to examine the structure of PRC2 complex36; however, crystal structures are not yet available. The complex is larger than 230 kDa; thus, it is challenging to crystallize. Nevertheless, the study of individual subunits can contribute to the study of the entire complex structure.
Role of EED/ESC
Of the five subunits of PRC2, the crystal structures of only EED and RbAp48 have been determined. The EED subunit consists of a WD-repeat domain that folds into a seven-bladed β-propeller (Figure 4A–4C)37,38,39 with an 80-residue N-terminus that is predicted to be unstructured. The WD-40 domain is found in functionally diverse proteins, with a doughnut-like structure that normally provides a scaffold for interactions with partner proteins or effectors3. Thus far, structural and functional studies have focused on three modules of EED or ESC (the homolog of EED in Drosophila). The first module is the interface on the bottom of the β-propeller, which binds an N-terminal fragment of EZH2 (residues 39–68)37,40. The second module is the pocket on the top of the β-propeller, which binds H3K27me3 or other histone marks38,39. The last module is the N-terminus, which is the unstructured region ahead of the WD-40 domain that can bind the histone-fold domain of H341.
Crystal structures of EED complexes or NURF-55 complexes. (A) The EZH2 N-terminal peptide (residues 39-68) binding to the bottom of the EED WD domain (PDB code: 2QXV). (B) H3K27me3 binding to an aromatic cage (top) on EED (PDB code: 3IIW). (C) Side view of the EED structure for comparing the binding of the EZH2 peptide and H3K27me3. (D) SU(Z)12 N-terminal peptide (residues 79-91) binding to the S/H-site of NURF-55 (PDB code: 2YBA). (E) Histone H31-19 peptide binding to the C-site of NURF-55 (PDB code: 2YB8).
Regarding the N-terminal EZH2 fragment and the EED WD-40 domain, GST pull-down assays have shown that a longer N-terminal fragment (residues 1–259) of EZH2 can form a stable complex with full-length EED; however, only the shorter peptide (residues 39–68) can be crystallized for structure determination37. Structural and mutagenesis studies indicate that the peptide-binding groove is lined mainly with hydrophobic residues on the bottom; however, the central region on the top of the WD-repeat domain of Drosophila ESC is more likely involved in interacting with other proteins42. Indeed, the top of the β-propeller of EED can specifically bind to histone tails carrying trimethyl-lysine residues such as H3K27me3 and H3K9me3 that are associated with repressive chromatin marks, which leads to the allosteric activation of the methyltransferase activity of PRC238 (Figure 4B and 4C).
In addition, both in vitro and in vivo data indicate that EED, as a non-catalytic subunit, makes a crucial contribution to PRC2 methyltransferase activity through its interaction with the N-terminal residues of EZH243. This finding is supported by several pieces of evidence. First, the ESC mutations M236K and V289M, which are located on the surface loop and mediate direct contact between ESC and E(Z) (the EZH2 homolog in Drosophila), can significantly reduce methyltransferase activity. In addition, ESC cannot bind to nucleosomes by itself, and it has little effect on nucleosome binding when added into the NURF-55/SU(Z)12/E(Z) complex [NURF-55 and SU(Z)12 are the homologs of human RbAP48 and SUZ12, respectively]. This finding is also supported by evolutionary data because every organism examined — from plants to flies to humans — has an ESC homolog. It has been reported43 that 28 residues within the ESC surface loops that are implicated in EZH2 binding are identical among these organisms, reflecting the functional requirement of EED/ESC as a partner in histone methyltransferase (HMTase) activity.
It is also important to note that the multiple EED isoforms expressed in human cells differ with respect to the length of their N-terminal tails and can be generated by using alternative start codons in the same EED mRNA44. Importantly, the incorporation of particular EED isoforms in the PRC2 complex can alter the enzyme specificity; for example, certain complexes may methylate H1K26 in addition to H3K27. Thus, it appears that the EED subunit can influence both the catalytic efficiency and the preferred lysine substrate.
Functional contribution of SUZ12
SUZ12 is also required for the HMTase activity of the PRC2 complex; however, it is unclear how it contributes to the overall molecular machine. It is possible that SUZ12 may directly interact with EZH2 and/or mediate nucleosome binding. SUZ12 has a highly conserved VRN2-EMF2-FIS2-SUZ12 (VEFS) domain at its C-terminus. Deletion of the entire VEFS domain can prevent assembly of the PRC2 complex in fly and mammalian systems because SUZ12-EZH2 binding is disrupted45. Thus, the conserved function of this domain is interaction with EZH2. In addition, mutations in this domain, such as D546A and E550A, can preserve full complex assembly but reduce HMTase levels. Further, in the fly system, the C-terminus of SU(Z)12 harboring the VEFS domain is the minimal domain required for HMTase activity and for inhibition of active marks such as H3K4 and H3K3646.
SUZ12 is also necessary for gene silencing by the PRC2 complex47, and one role of SUZ12 is to mediate nucleosome binding, although it cannot bind to a nucleosome by itself48. It has been suggested that binding of the chromatin activation marks H3K4me3, H3K36me2, and H3K36me3 can inhibit PRC2 activity if they are located in cis on the same histone molecule containing the target lysine H3K27, and SU(Z)12 is responsible for mediating this inhibition in conjunction with the E(Z) SET domain46. Because this effect can be reversed by the addition of an H3K27me3-containing peptide, it has been thought that PRC2 can simultaneously integrate inhibitory and activating chromatin marks, tuning its enzymatic activity based on the surrounding chromatin state36,46. The minimal PRC2 subcomplex that retains both HMTase activity and H3K4me3/H3K36me2/3 inhibition has been reported to contain EZH2 and two activity-controlling elements of PRC2, namely the WD40 domain of EED and the VEFS domain of SUZ1236,38,46.
Possible role of RbAp48
RbAp48 and its Drosophila homolog NURF-55 contribute only minimally to the HMTase activity of the PRC2 complex43,47; another subunit containing a WD-40 domain binds to the N-terminus of SUZ1246,49. Unlike the E(Z), ESC, and SU(Z12) subunits, which only exist in the PRC2 complex, NURF-55 has been found in diverse chromatin-modifying complexes, such as chromatin assembly factor 1, NURF, and nucleosome remodeling and deacetylase complexes49,50,51. Because RbAp48/NURF-55 is not essential for robust PRC2 methyltransferase activity47, there are long-standing questions regarding what major role RbAp48/NURF-55 plays in the PRC2 complex.
The crystal structures of NURF-55 in complex with an H3 peptide (1–19 aa) and the N-terminal peptide of SUZ12 (79–93 aa) have been determined46 (Figure 4D, 4E). These two peptides bind to different sites on NURF-55, and the two binding interactions are not interdependent. Based on the structures and biochemical experiments, it has been proposed that NURF-55–SUZ12 plays a role in the sequestration and release of histone H346. H3K4me3 inhibition can be induced by the release of the histone H3 tail, triggering the allosteric inhibition of PRC2 activity. However, some other active marks, such as H3K9me3- and H3R2me-modified tails, are not expected to interfere with PRC2 regulation because they have no effect on PRC2 activity in vitro. In addition, the interactions between NURF-55 and other active marks might play a subtle role in positioning the complex correctly on nucleosomes.
AEBP2 acts as a co-factor of the PRC2 complex
AEBP2 acts as a co-factor that can stably interact with other subunits of the PRC2 complex, and it is required for optimal enzymatic activity47. Cross-linking data have shown that the central region of AEBP2 can interact with the N- and C-termini of EZH2 and EED as well as the C-terminus of SUZ12, while the C-terminus of AEBP2 interacts with the central portions of both SUZ12 and RbAp48. These data suggest an important role for AEBP2 in coordinating different PRC2 subunits and stabilizing the PRC2 complex36.
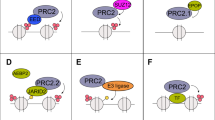
The overall architecture of the PRC2 complex has been proposed based on biochemical data and electron microscopy analysis. In this architecture, a structural core consisting of EZH2, the WD40 domain of EED, and the VEFS domain of SUZ12 is thought to have HMTase activity, and the ability of these domains to regulate PRC2 activity could be explained by their physical interaction with the SET domain of EZH2. At the opposite end of the core module, both the VEFS domain of SUZ12 and the SANT and SET domains of EZH2 interact together with two Zn fingers from AEBP2 and SUZ12. The position of the SUZ12 VEFS domain with respect to the EZH2 SET domain suggests how the inhibition of H3K4me3 and H3K36me2/3 via SUZ12 could be transferred to EZH2, thereby regulating its methyltransferase activity. The close proximity of these elements could also explain why the gene activation marks must be on the tail being modified to block methyltransferase activity.
In summary, the mechanism of action and allosteric regulation of PRC2 during gene silencing have been proposed based on the architecture of PRC2 and biochemical data concerning the interactions among the subunits. When the chromatin state is compact and repressed, existing H3K27me3 marks are recognized by EED. The SANT domains send a signal to the SET domain indicating that EED binding has occurred, which increases the methyltransferase activity of EZH2 and strengthens chromatin compaction. When the chromatin state is open and actively transcribed, H3K4me3 and H3K36me2/3 are recognized by the VESF domain of SUZ12 and transferred to EZH2, leading to allosteric regulation that blocks the enzymatic activity of EZH236.
Methyltransferase SET domain
The histone methyltransferases form a large family that catalyzes methyl group transfer from a universal methyl donor, S-adenosyl-L-methionine (SAM; also known as AdoMet), to the nitrogen atom of the lysine side chain, forming S-adenosyl-L-homocysteine (SAH; also known as AdoHcy) as a product2. The catalytic domain is quite conserved among the methyltransferases and contains approximately 130 amino acids. Although EZH2 contains this conserved SET domain, it cannot perform the catalytic function without two other subunits of the PRC2 complex.
A sequence alignment of various SET-domain-containing methyltransferases is shown in Figure 5, and some related structures are shown in Figure 6. Of these structures, the proteins Clr452, Dim553, SET7/954, and G9a55 adopt a typical fold composed of a conserved SET domain with variable insertions between its two non-contiguous regions, known as SET-N and SET-C. The SET domains are flanked by a pre-SET region (cysteine-rich, also referred to as a CXC domain) and a post-SET region, and the SET domains are characterized by canonical features such as a knot-like region adjacent to the catalytic site, a distinct co-factor binding site, and a substrate binding pocket56. The catalytic site is assumed to contain N241, H242, and Y283 (in Dim5 residue numbering), as they are invariant among these SET domains (Figure 5) and they are located in the correct position according to structural analysis57. Y283 functions as a general base to deprotonate the lysine substrate before methyl transfer; however, it is unclear how the quaternary nitrogen atom is deprotonated to generate a neutral amine methyl acceptor when both the tyrosine and lysine are deprotonated at a physiological pH owing to their high pKa values. One hypothesis based on molecular dynamics simulation is that the binding of the cofactor SAM and the substrates creates a water shuttle, which can facilitate the deprotonation of the lysine side chain58.
Structure-based sequence alignment of selected SET domains containing histone methyltransferase. The alignment includes Clr4 (NP_595186), Dim5 (AAL35215), SUV39h1 (NP_003164), G9a (S30385), EZH2 (NP_004447), SET7/9 (NP_085151), SET8 (NP_065115), and vSET (AAC96946). The conserved sequence are highlighted with green boxes. Abbreviations: Hs, Homo sapiens; Nc, Neurospora crassa; Sp, Schizosaccharomyces pombe. The catalytic sites are indicated with star, and mutations of Y641 and A677 are indicated with triangles.
Structures of the protein methyltransferase SET domain. Zinc atoms are shown in grey spheres. (A) Clr4_Sp (PDB code: 1MVX). (B) Dim5_Nc complexed with H3K9 (pink sticks) and SAH (yellow sticks) (PDB code: 1PEG). (C) SET7/9_Hs complexed with H3K9 peptide and SAH (PDB code: 1O9S). (D) G9a-like protein methyltransferase complexed with BIX-01294 (cyan sticks) and SAH (PDB code: 3FPD). (E) SET8 complex with SAH (PDB code: 1ZKK). (F) vSET from virus complexed with peptide and SAH (PDB code: 2G46).
The pre-SET domain is a cysteine-rich region that can form a triangular Zn3Cys9 zinc cluster and play an important role in the catalytic reaction52,53, whereas the structure of the post-SET domain is variable. Dim5, G9a, GLP, and SUV39H255 include a ZnCys motif (Figure 5), which contributes to substrate binding. SET7/9 does not have a Cys-rich domain, and it relies on the C-terminal residues of the SET domain for the formation of a lysine channel by packing an α-helix onto the active site, rather than a metal center (such as a zinc cluster)56. Based on these structures, the conformational states of the post-SET domain can be divided into three types according to the co-factor and/or substrate binding: 1) a flexible (or even disordered) state without co-factor or substrate binding; 2) a loose conformation with SAH but no substrate binding; and 3) a relatively rigid conformation with both SAH and substrate binding. These states are most likely in a dynamic equilibrium and can shift to the last state upon co-factor and substrate binding55.
The vSET domain from Paramecium bursaria chlorella virus 1 can also methylate H3K27, and it is the only structure determined so far that contains SAH and the H3K27 peptide59. This solution structure forms a dimer and has no post-SET domain, which distinguishes it from the structures mentioned above. This complex structure explains the basis of the substrate specificity of vSET for H3 at K27, but not at K9, through recognition of the APA motif (H3 residues 29–31), which only exists in H3K27. In addition, Y105, which is conserved in other methyltransferases, is thought to facilitate methyl transfer from SAM to H3K27 by aligning their intermolecular interactions at the substrate lysine access channel in the enzyme. These data stand in contrast to what is known about EZH2, which does not need to form a dimer to perform the catalytic reaction, but instead requires two other subunits of the PRC2 complex.
Wild-type EZH2 has the greatest catalytic activity for monomethylation of H3K27, although its mutants (such as Y641F and A677G) perform the subsequent reactions with high efficiency. Other SET-domain protein methyltransferases (PMTs), such as SET7/9, produce only monomethylated products after one round of catalysis, whereas G9a and GLP are mono- and dimethylases, and SUV39h2 can di- and trimethylate a monomethylated substrate55. According to the crystal structures, the difference between single-turnover and multiple-turnover SET domain enzymes results from the degree of steric crowding and hydrogen bonding patterns in the lysine-binding channel of these enzymes60,61. One or two aromatic residues within the lysine-binding pocket are thought to play important roles in this procedure. For example, on one side, Dim5 can trimethylate the substrate H3K9 with the aid of an aromatic phenylalanine residue (F281), while the Dim5 mutant F281Y can catalyze monomethylation. It has also been shown that the Y1067F mutant of G9a is able to trimethylate H3K9, whereas wild-type G9a catalyzes mono- and dimethylation55. Thus, a “tyrosine-phenylalanine switch” appears to be a general determinant of product specificity among the SET domain-containing methyltransferases2,62.
In summary, the characterization of these methyltransferase structures — especially the structures of their active sites — provides an opportunity for the development of inhibitors that can compete with either SAM or lysine. It may also be possible to develop highly selective inhibitors according to the differences in these binding pockets.
Progress in drug discovery
DZNep as an indirect inhibitor
Over the past few years, several potent inhibitors of EZH2 have been discovered. Among these inhibitors, 3-deazaneplanocin A (DZNep), a S-adenosylhomocysteine hydrolase inhibitor (Figure 7), depletes EZH2 and the associated H3K27me3 and can induce apoptosis in breast and colon cancer cells63. DZNep interferes with S-adenosylmethionine and SAH metabolism and can indirectly inhibit the methylation reaction. DZNep-induced apoptosis is partially related to its ability to inhibit the PRC2 pathway, although the exact mechanism has not yet been elucidated. In addition, it has been reported that DZNep is synergistic with histone deacetylase inhibitors and DNA methyltransferase inhibitors in the activation of silenced genes64. DZNep has minimal toxicity in vivo as an antiviral compound65, and together with its importance in cancer epigenetic pathways, it may be a promising drug candidate for anti-cancer treatment.
PRC2 subunit composition and modes of inhibition. Three types of inhibitors are indicated: SAM competitive inhibitors, SAH, and DZNep as an SAH hydrolase inhibitor.
SAM-competitive inhibitor
SAH, a universal product of SAM hydrolysis, has a Ki value of 75 μmol/L against EZH2 and an IC50 value of 0.1–20 μmol/L among PMTs. SAH-dependent methylation is involved in many cellular processes66; consequently, SAH has low selectivity against other methyltransferases63. Sinefungin is a nonspecific SAM analog that has similar potency67. Because neither of these compounds shows specificity against EZH2, significant efforts have been made over the past few years to obtain compounds that are potent and highly selective for PRC2 (Table 2)68,69,70,71.
To identify inhibitors of EZH2 methyltransferase activity, high-throughput biochemical screening experiments have been performed with the PRC2 complex. Several potent inhibitors were identified with Ki values in the low nanomolar range. Although the structure of the EZH2 active site has not yet been determined, the conserved SET domain architecture predicts two essential binding pockets: one for the SAM methyl donor and another for the Lys27 substrate. Because more than 50 SET domain proteins have been identified in humans thus far, the selectivity of the inhibitors is crucial for minimizing off-target effects2.
At the end of 2012, several SAM-competitive inhibitors were announced. The compound EPZ005687 has a Ki value of 24 nmol/L and is over 500-fold more selective for EZH2 versus 15 other PMTs and 50-fold more selective for EZH2 versus the closely related enzyme EZH168. EPZ005687 can also inhibit H3K27 methylation by the EZH2 mutants Y641 and A677, and it has been shown to selectively kill lymphoma cells that are heterozygous for one of these EZH2 mutations68.
EI1, another inhibitor of EZH2, was developed by Novartis71 and shows very good selectivity with a low Ki value (approximately 13 nmol/L). Loss of the H3K27 methylation function and activation of PRC2 target genes have been observed in EI1-treated cells. EI1 is equally active against both wild type and the Y641 mutant form of EZH2, and the inhibition of the EZH2 Y641 mutant in B-cell lymphomas leads to decreased proliferation, cell cycle arrest, and apoptosis71.
The most potent inhibitor of EZH2 thus far is GSK126, which has a Ki of 0.5–3 nmol/L70. The selectivity of GSK126 for EZH2 is more than 1000-fold higher than its selectivity for 20 other human methyltransferases containing SET or non-SET domains, and it is over 150-fold more selective for EZH2 than for EZH1. GSK126 effectively inhibits the proliferation of EZH2 mutants in DLBCL cell lines. More importantly, at the animal level, GSK126 markedly inhibits the growth of EZH2-mutant DLBCL xenografts in mice, which was the first animal model for studying the antitumor effects of EZH2. Thus, GSK126 provides a valuable means for evaluating whether EZH2 activity is required for the survival of tumors in which EZH2 overexpression has been linked to poor prognosis. Taken together, these data suggest that pharmacological inhibition of EZH2 activity may be a feasible strategy for treating DLBCLs and non-indolent follicular lymphomas that harbor activating mutations in EZH2.
UNC1999, an analogue of GSK126, is the first orally bioavailable inhibitor that has high in vitro potency against wild type and mutant EZH2 over a broad range of epigenetic and non-epigenetic targets. However, UNC1999 shows less selectivity for EZH1 than the inhibitors mentioned above. Because no crystal structure is available, it is not clear how structural changes contribute to the high selectivity of GSK126 (over 150-fold) versus UNC1999 (approximately 10-fold) for EZH2/EZH1. UNC1999 potently reduced H3K27me3 levels in cells (IC50<50 nmol/L) and selectively killed DLBCL cell lines harboring the Y641N mutation69. Because it is orally bioavailable in mice, UNC1999 could function as a chemical probe for investigating the role of EZH2 in chronic animal studies.
Many competitive inhibitors bind to the SAM pocket; however, a large number of inhibitors can also bind to the protein-substrate binding site, engaging recognition elements within the amino acid channel. BIX-01294 is one such inhibitor (Table 2 and Figure 6) and was found to be a potent inhibitor of G9a with an IC50 of approximately 1.0 μmol/L72.
The SAM competitive inhibitors were also tested against vSET, a viral lysine methyltransferase that is the smallest protein unit capable of catalyzing H3K27 methylation. The sequence identity between the SET domains of EZH2 and vSET is only 23%, and there are significant differences in the active sites of these enzymes73. In addition, vSET folds into a tertiary structure as a homodimer that can methylate H3K2774. Because EZH2 requires at least other two subunits (EED and SUZ12) for its activity, several key differences exist between these two enzymes, including the size of the protein, overall structure, and substrate recognition. Thus, it is not surprising that highly potent inhibitors, such as EPZ005687 and GSK126, are not able to inhibit vSET73. The data clearly reflect significant structural differences between the two enzymes with respect to their SAM binding pockets, which precludes the use of vSET as a meaningful tool for structure-based EZH2 inhibitor design.
In summary, the function of PRC2 as transcriptional repressor or gene activator has been explored as the basis for drug discovery. Although no compounds are currently approved for treatment or clinical trials, much effort has been made to develop EZH2 methyltransferase inhibitors. Because EZH2 is an attractive anti-tumor target, additional research aimed at the discovery and design of EZH2 inhibitors is warranted.
Future directions
Over the past few years, significant progress has been made in the study of the anti-tumor mechanism of PRC2 and the design of inhibitors against this target. Mechanistic studies have shown that PRC2 catalyzes H3K27 methylation and contains a recognition site for binding to this modification. Additionally, PRC2 also harbors a control module that triggers inhibition of this activity to prevent H3K27 trimethylation in transcriptionally active genes. PRC2 can thus integrate information provided by preexisting histone modifications to accurately tune its enzymatic activity within a particular chromatin context. Nevertheless, important questions still remain.
To fully reveal the functions of each subunit, the structure of the PRC2 complex must be determined; however, this is still a major challenge. At the very least, the core structure of PRC2 (ie, EZH2-EED-SUZ12) should be determined to better understand the interactions between the core subunits . However, the mechanisms of allosteric inhibition need to be addressed by protein dynamics studies (for example, molecular dynamics simulations) to determine how conformational changes could deliver functional effects between the PRC2 subunits. Furthermore, although it is still early in the development of methyltransferase inhibitors for clinical application, drug resistance and low selectivity are common issues of concern for all targeted cancer drugs. The ability to effectively combine the current anti-tumor agents, such as DNA methyltransferase inhibitors and histone deacetylase inhibitors, with EZH2 inhibitors will be a major clinical challenge in the future.
References
Pollock RM, Richon VM . Epigenetic approaches to cancer therapy. Drug Discov Today: Ther Strat 2009; 6: 71–9.
Copeland RA, Solomon ME, Richon VM . Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov 2009; 8: 724–32.
O'Meara MM, Simon JA . Inner workings and regulatory inputs that control Polycomb repressive complex 2. Chromosoma 2012; 121: 221–34.
Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298: 1039–43.
Simon JA, Lange CA . Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 2008; 647: 21–9.
Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 2005; 15: 942–7.
Montgomery ND, Yee D, Montgomery SA, Magnuson T . Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. J Mol Biol 2007; 374: 1145–57.
Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K . Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004; 23: 4061–71.
Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res 2007; 67: 10657–63.
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004; 431: 873–8.
Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell 2011; 20: 187–99.
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439: 871–4.
Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ . The drosophila polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 2001; 128: 275–86.
van der Vlag J, Otte AP . Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet 1999; 23: 474–8.
Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010; 42: 181–5.
Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A 2010; 107: 20980–5.
Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 2011; 117: 2451–9.
McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci U S A 2012; 109: 2989–94.
Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell 2011; 43: 798–810.
Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol 2007; 27: 5105–19.
Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012; 338: 1465–9.
Deb G, Thakur VS, Gupta S . Multifaceted role of EZH2 in breast and prostate tumorigenesis: epigenetics and beyond. Epigenetics 2013; 8: 464–76.
Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 2010; 116: 5247–55.
Bohrer LR, Chen S, Hallstrom TC, Huang H . Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: a potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology 2010; 151: 5136–45.
Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–9.
Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol 2006; 24: 268–73.
Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K . EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22: 5323–35.
Gonzalez ME, DuPrie ML, Krueger H, Merajver SD, Ventura AC, Toy KA, et al. Histone methyltransferase EZH2 induces Akt-dependent genomic instability and BRCA1 inhibition in breast cancer. Cancer Res 2011; 71: 2360–70.
Hervouet E, Cartron PF, Jouvenot M, Delage-Mourroux R . Epigenetic regulation of estrogen signaling in breast cancer. Epigenetics 2013; 8: 237–45.
Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A 2003; 100: 11606–11.
Morin RD, Mendez–Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011; 476: 298–303.
van Kemenade FJ, Raaphorst FM, Blokzijl T, Fieret E, Hamer KM, Satijn DP, et al. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood 2001; 97: 3896–901.
Zhao X, Lwin T, Zhang X, Huang A, Wang J, Marquez VE, et al. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia 2013; 27: 2341–50.
Hubaux R, Thu KL, Coe BP, Macaulay C, Lam S, Lam WL . EZH2 promotes E2F-Driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol 2013; 8: 1102–6.
Zhao JC, Yu J, Runkle C, Wu L, Hu M, Wu D, et al. Cooperation between polycomb and androgen receptor during oncogenic transformation. Genome Res 2012; 22: 322–31.
Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E . Molecular architecture of human polycomb repressive complex 2. eLife 2012; 1: e 00005.
Han Z, Xing X, Hu M, Zhang Y, Liu P, Chai J . Structural basis of EZH2 recognition by EED. Structure 2007; 15: 1306–15.
Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009; 461: 762–7.
Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc Natl Acad Sci U S A 2010; 107: 19266–71.
Wu Z, Lee ST, Qiao Y, Li Z, Lee PL, Lee YJ, et al. Polycomb protein EZH2 regulates cancer cell fate decision in response to DNA damage. Cell Death Differ 2011; 18: 1771–9.
Tie F, Stratton CA, Kurzhals RL, Harte PJ . The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol Cell Biol 2007; 27: 2014–26.
Tie F, Furuyama T, Harte PJ . The Drosophila Polycomb group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development 1998; 125: 3483–96.
Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA . Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol 2005; 25: 6857–68.
Kuzmichev A, Jenuwein T, Tempst P, Reinberg D . Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 2004; 14: 183–93.
Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T . Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2. J Biol Chem 2004; 279: 401–6.
Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 2011; 42: 330–41.
Cao R, Zhang Y . SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 2004; 15: 57–67.
Nekrasov M, Wild B, Muller J . Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep 2005; 6: 348–53.
Martinez-Balbas MA, Tsukiyama T, Gdula D, Wu C . Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci U S A 1998; 95: 132–7.
Lejon S, Thong SY, Murthy A, AlQarni S, Murzina NV, Blobel GA, et al. Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48.FOG-1 complex. J Biol Chem 2011; 286: 1196–203.
Verreault A, Kaufman PD, Kobayashi R, Stillman B . Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 1996; 87: 95–104.
Min J, Zhang X, Cheng X, Grewal SI, Xu RM . Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol 2002; 9: 828–32.
Zhang X, Tamaru H, Khan SI, Horton JR, Keefe LJ, Selker EU, et al. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 2002; 111: 117–27.
Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 2003; 421: 652–6.
Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, et al. Structural biology of human H3K9 methyltransferases. PLoS One 2010; 5: e8570.
Xiao B, Wilson JR, Gamblin SJ . SET domains and histone methylation. Curr Opin Struct Biol 2003; 13: 699–705.
Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell 2003; 12: 177–85.
Zhang X, Bruice TC . Enzymatic mechanism and product specificity of SET-domain protein lysine methyltransferases. Proc Natl Acad Sci U S A 2008; 105: 5728–32.
Qian C, Wang X, Manzur K, Sachchidanand, Farooq A, Zeng L, et al. Structural insights of the specificity and catalysis of a viral histone H3 lysine 27 methyltransferase. J Mol Biol 2006; 359: 86–96.
Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem 2005; 280: 5563–70.
Smith BC, Denu JM . Chemical mechanisms of histone lysine and arginine modifications. Biochim Biophys Acta 2009; 1789: 45–57.
Cheng X, Collins RE, Zhang X . Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct 2005; 34: 267–94.
Tan J, Yang XJ, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007; 21: 1050–63.
Sun F, Chan E, Wu Z, Yang X, Marquez VE, Yu Q . Combinatorial pharmacologic approaches target EZH2-mediated gene repression in breast cancer cells. Mol Cancer Ther 2009; 8: 3191–202.
Bray M, Driscoll J, Huggins JW . Treatment of lethal Ebola virus infection in mice with a single dose of an S-adenosyl-L-homocysteine hydrolase inhibitor. Antiviral Res 2000; 45: 135–47.
Chiang PK . Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacol Ther 1998; 77: 115–34.
Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC . Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J Biol Chem 2006; 281: 19280–7.
Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 2012; 8: 890–6.
Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem Biol 2013; 8: 1324–34.
McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012; 492: 108–12.
Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A 2012; 109: 21360–5.
Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol 2009; 16: 312–7.
Swalm BM, Hallenbeck KK, Majer CR, Jin L, Scott MP, Moyer MP, et al. Convergent evolution of chromatin modification by structurally distinct enzymes: comparative enzymology of histone H3 Lys27 methylation by human polycomb repressive complex 2 and vSET. Biochem J 2013; 453: 241–7.
Wei H, Zhou MM . Dimerization of a viral SET protein endows its function. Proc Natl Acad Sci U S A 2010; 107: 18433–8.
Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A . Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol 2005; 1: 143–5.
Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 2007; 25: 473–81.
Acknowledgements
We thank David NADZIEJKA for scientific editing and critical comments. This work was supported in part by the Jay and Betty Van Andel Foundation, Amway (USA), the National Natural Science Foundation of China (NSFC 81123004), and Ministry of Science and Technology (China) grants 2012CB910403 and 2013CB910601.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Tan, Jz., Yan, Y., Wang, Xx. et al. EZH2: biology, disease, and structure-based drug discovery. Acta Pharmacol Sin 35, 161–174 (2014). https://doi.org/10.1038/aps.2013.161
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2013.161