Abstract
Background:
Neurofibromatosis type 1 is one of the most common familial diseases, the hallmark of which is the development of multiple neurofibromas. These are benign nerve sheath tumours, which can transform into malignant peripheral nerve sheath tumours (MPNST).
Methods:
The aim of this study was to identify differentially expressed microRNA (miRNA) in neurofibromas and MPNST obtained from patients with neurofibromatosis type 1 using microarray analysis. Differential expression was validated by reverse transcription quantitative–PCR, and functional studies were performed after transfection of miRNA oligonucleotide mimics into MPNST cells.
Results:
Sixteen miRNA were significantly differentially expressed in MPNST compared with NF, and of these fourteen were downregulated in MPNST: these included miR-30e*, miR-29c*, miR-29c, miR-340*, miR-30c, miR-139-5p, miR-195, miR-151-5p, miR-342-5p, miR-146a, miR-150, miR-223, let-7 a and let-7 g with a false discovery rate of q=8.48E-03 for the least significant miRNA. In contrast, miR-210 and miR-339-5p were upregulated in MPNST compared with neurofibromas. Prediction softwares/algorithms identified a list of genes targeted by miR-29c including extracellular matrix genes and matrix metalloproteinase (MMP)-2, all of which are reported to be involved in cell migration and invasion. Functional studies in a MPNST cell line, sNF96.2, using a mimic of the mature miR-29c showed reduced invasion, whereas there was no change in proliferation. Zymography of the manipulated cells showed that MMP2 activity was also reduced when miR-29c expression was forced in sNF96.2.
Conclusion:
We provide evidence that reduction of miR-29c has a pivotal role in the progression of nerve sheath tumours and results by increasing the invasive/migratory properties of nerve sheath tumours.
Similar content being viewed by others
Main
Neurofibromatosis type 1 is caused by biallelic inactivation of the tumour suppressor gene neurofibromin 1 located in 17q11.2 (Gutmann et al, 1997), a common familial disease affecting 1 in ∼3000 of the population, the hallmark of which is the development of multiple neurofibromas. These are benign tumours which derive from Schwann cells. Approximately 10% of patients with neurofibromatosis type 1 develop a malignant peripheral nerve sheath tumour (MPNST), a tumour, which is associated with a high risk of mortality (King et al, 2000; Kleihues and Cavenee, 2000; Evans et al, 2002). Surgery is the standard treatment for MPNST, with chemotherapy offering little benefit (Kroep et al, 2011). The overall 5-year survival for individuals with malignant disease ranges from 40 to 75% for the sporadic MPNST to 21% for NF1 MPNST patients (Evans et al, 2002; Porter et al, 2009). Hence, understanding how neurofibromas transform to MPNST would be potentially beneficial as it may offer the opportunity to provide an early diagnosis and identify therapeutic targets.
Gene expression microarrays (GEM) and more recently a whole-genome comparative methylome analysis have been performed on neurofibromas and MNPST from patients with neurofibromatosis type 1 (Miller et al, 2006; Feber et al, 2011). The former analysis shows an overall downregulation of genes in MPNST compared with the benign counterpart, consistent with our finding of a large number of genes being hypermethylated in MPNST (Feber et al, 2011). The relatively few genes expressed at higher levels in MPNSTs compared with neurofibromas included a number of genes involved in cell proliferation and in tumour metastasis (Subramanian et al, 2010) including collagen type VIa3, and collagen type VIIa1, and related genes including survivin, and tenascin C (Karube et al, 2006; Subramanian et al, 2010).
MicroRNA (miRNA) are small, processed, non-coding RNAs molecules ∼22 nucleotides in length that regulate gene function via mRNA degradation, and translation inhibition or chromatin-based silencing mechanisms (Bartel, 2004). They influence a wide variety of physiological and pathological processes, including tumour biology where they mediate their effect as either tumour suppressor genes or oncogenes (Pasquinelli et al, 2005; Calin and Croce, 2006; Esquela-Kerscher and Slack, 2006; Garzon et al, 2006; Di and Croce, 2010). MicroRNAs profiling of benign and MPNSTs has shown a relative downregulation of miR-34a in most MPNSTs compared with neurofibromas from a population of patients with and without neurofibromatosis type 1 (Subramanian et al, 2010). The aim of this study was to identify additional differentially expressed miRNA in neurofibromas and MPNST obtained from patients with neurofibromatosis type 1 for the purpose of understanding the mechanism by which neurofibromas transform into MPNSTs.
Materials and methods
Clinical samples
This study was approved by the Central Office for Research Ethics Committee (REC 07/Q0506/8). The material was obtained from the histopathology department of The Royal National Orthopaedic Hospital and three cases from Department of Clinical Genetics were kindly provided by Dr Fredrik Mertens, Lund University Hospital, Lund, Sweden. The pathology slides were reviewed, diagnosed and graded according to WHO guidelines (Kleihues and Cavenee, 2000; Fletcher et al, 2002). The material for the study included frozen material obtained from 10 neurofibromas (four female, six male) and 10 high-grade MPNSTs (five female, five male) from patients with neurofibromatosis type 1. Analysis of the same 10 neurofibromas and 10 MPNST individually by reverse transcription quantitative–PCR (RT–qPCR) for neurofibromin 1 expression confirmed very-low expression of the gene in both the neurofibromas and MPNST cases compared with HEK293T cells, which are neurofibromin 1 wildtype (WT) (Supplementary Figure S1A, Supplementary Table S1 for primers sequences). This finding along with loss of heterozygosity of the neurofibromin 1 locus, as recently reported by SNP genotyping analysis on the pooled samples from the same 10 neurofibromas and 10 MPNST (Feber et al, 2011), is consistent with functionally biallelic inactivation of neurofibromin 1 in these tumours (Legius et al, 1993; Colman et al, 1995).
An additional 116 tumours (56 neurofibromas and 60 MPNST) obtained from patients with and without neurofibromatosis type 1 were used for validation purposes.
Cell culture
The human MPNST cell line sNF96.2, was kindly provided by Dr Abhijit Guha (The hospital for Sick Children MaRS, Toronto, Ontario, Canada). We confirmed the reported germline mutation in exon 21 (3683delC) and loss of the remaining WT allele in the cell line before use in our study compared with HT1080 WT for neurofibromin 1 (Supplementary Figure 1B, Supplementary Table S1 for primers sequences) (Perrin et al, 2007). The cell line was grown in DMEM supplemented with 10% foetal bovine serum, and 1% penicillin and streptomycin at 37 °C in a 5% CO2 incubator.
Nucleic acid isolation
Between 20 and 30 frozen sections (20 μm) from sampled neoplasms comprising at least 60% tumour cells were used for nucleic acid extraction. Total RNA, including miRNA was extracted using the miRNAeasy kit (Qiagen Ltd, Crawley, West Sussex, UK).
Agilent Human miRNA Microarray V2 and data analysis
The small RNA fraction was assessed using the Agilent small RNA Kit Lab-on-Chip (Agilent Technologies UK Ltd, Wokingham, UK). Quality and integrity of the RNA fraction, between 6 and 150 nucleotides, was quantified using the Agilent 2100 bioanalyser (Agilent Technologies). Hundred nanogram of total RNA per sample was labelled and hybridised to the Agilent Human miRNA microarray V2 (Agilent Technologies).
Microarray slides were scanned using an Agilent microarray scanner G2 505B (Agilent Technologies) and microarray images were automatically analysed using Feature extractionTM software, version 9.5.1.1 (Agilent Technologies). microRNA analysis was carried out using Bioconductor packages for the R statistical programming language (Gentleman et al, 2004). The median (gMedianSignal) was background subtracted (gBGUsed) and then the arrays normalised to each other using the ‘normexp’; function (Ritchie et al, 2007) of the limma package. The limma package was also used for differential expression and multiple testing was controlled using the false discovery rate (q-value).
Quantitation of miRNA by TaqMan assay
MicroRNA expression in each sample, and the cell line, was measured using TaqMan miRNA assays technology (Applied Biosystems Inc., Life Technologies, Foster City, CA, USA) on a Mastercycler realplex Eppendorf detection system (Eppendorf, Histon, Cambs., UK). This RT–qPCR method specifically detects mature miRNA, but not precursor miRNA (pre and pri-miRNA). The TaqMan assay was performed using the same amount of total RNA (5 ng) that was transcribed into cDNA (Applied Biosystems Inc.), and Ct-values were recorded. The mean of three independent analyses was calculated and an internal control small RNA, RNU66 was used to normalise the miRNA expression data for each sample (Applied Biosystems).
Detection of the target genes by RT–qPCR
Approximately 150–200 ng of the total RNA extracted from each sample was reverse transcribed using Superscript III First-Strand Synthesis kit (Invitrogen Ltd/Life Technologies Ltd, Paisley, Scotland, UK). Specific gene primers were designed with Primer 3 (http://frodo.wi.mit.edu/primer3/) and real-time PCR was performed with a SYBR green mix from Applied Biosystems (Supplementary Table S1 for primer sequences). The average of three independent analyses for each target gene and sample was calculated and was normalised to the endogenous reference control gene, GAPDH.
Mimic miR-29c transient transfection
The MPNST cell line, sNF96.2, was transfected in six-well plates using oligofectamine (Invitrogen) giving a final concentration of 100 nM of miRNA mimics (Dharmacon Inc., Lafayette, CO, USA). Cells were harvested after 24 h, 48 h, and 72 h following exposure to the mimics, after which total RNA including miRNA were extracted (Qiagen).
Functional studies
Migration assay
After 2 days of transfection with miR-29c, and scrambled-miR, sNF96.2 cells were harvested and ∼25 000 cells were plated in transwell plates with a polycarbonate membrane (8.5 mm diameter, 8.0 μm pore size) (Corning distributed by Appleton Woods, Birmingham, UK) and incubated for 6 h at 37 °C. Cells were washed with PBS and fixed with 4% formaldehyde, permeabilised with 0.1% Triton X-100 and stained with GelRed (Cambridge Bioscience, Cambridge, Cambs., UK). The number of cells that migrated across the filters was counted in 4 high-power fields per experiment, and the values averaged.
Scratch/wound healing assay
Two days after transfection with miR-29c and scrambled miRNA, the sNF96.2 cells were harvested and re-plated in six-well plates. On reaching confluency, a scratch was made through the central axis of the plate using a 10 μl pipette tip. The live-cell imaging system, IncuCyte (Essen BioScience Ltd, Herts., UK) was used to monitor the migration of cells across the scratch for 60 h.
Gelatin zymography
Two days after siRNA transfection of sNF96.2 cells the medium was changed to serum-free medium (350 μl (Optimem; Invitrogen Ltd) for overnight incubation after which the supernatant was collected and spun to remove floating cells. At the same time, the cells were lysed using lysis buffer (0.1% Triton x-100 in 0.05 Trizma base, 0.2 M NaCl, 0.005 CaCl2) in non-reduced conditions (no heating and no reducing reagents). The supernatants and cell extracts were separated using 10% acrylamide gels containing 0.1% gelatin type A from porcine skin (Sigma-Aldrich, Dorset, UK) with non-reducing conditions. The gels were incubated in a 2.5% Triton X-100 solution at room temperature with gentle agitation for 1 h and then soaked in reaction buffer (50 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 10 mM CaCl2) at 37 °C overnight. The gels were stained with 0.5% Coomassie Blue R250 in 30% methanol/10% in H2O for 1 h and then de-stained (staining solution without Coomassie Blue) for 0.5 h after which they were fixed in 0.2% glacial acetic acid. Zones of gelatinolytic activity were revealed by negative staining.
Cell viability assay
sNF96.2 cells were plated in triplicate at 25 000 cells per well in 96-well plates, and analysed at four time points, over 3 days. Cell proliferation was assessed using the MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2 H-tetrazolium, inner salt) assay (CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay; Promega, Southampton, UK). Cell viability was determined by absorbance measurements at 490 nm using a plate reader (Varioskan Flash, Thermo electrone corporation, Vantaa, Finland).
Growth curve
The rate of cellular proliferation was determined during and after transfection using a live-cell imaging system IncuCyte HD. The kinetic growth curves were calculated using the integrated confluence algorithm and the result was provided as the percentage of confluency.
Western blotting
Whole-cell lysate preparation and western blot analysis were performed in non-reduced conditions using Radio Immuno Precipitation Assay lysis buffer as described (Presneau et al, 2009). The primary antibodies used included: mouse monoclonal (CA-4001) matrix metalloproteinase (MMP)2 and rabbit monoclonal (EP1254) MMP9 (dilution 1 : 1000; incubation overnight at 4 °C, Abcam, Cambridge, Cambs., UK), MMP3 (dilution 1 : 50; incubation overnight at 4 °C, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and GAPDH mouse mAb (6C5) (dilution 1 : 5000; incubation 1 h at room temperature; Advanced Immunochemical Inc., Long Beach, CA, USA).
Statistical analysis
All experiments were performed as independent replicates and error bars correspond to the s.d. from the calculated mean. χ2-tests, Fisher exact tests, paired t-test or Wilcoxon tests were used, where applicable, to analyse differences between independent variants using R console (http://www.r-project.org/index.html) or GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA). Differences were considered significant if P<0.05.
Results
MicroRNA profiling identifies the miR-29 family as downregulated in MPNST
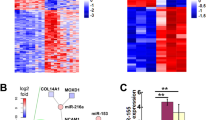
Multidimensional scaling of 10 NF and 9 MPNST (one failed to hybridise) following hybridisation on an Agilent miRNA microarray illustrated the similarity of expression between tumour groups (Figure 1). Translating these data into a two-dimensional plot demonstrated that apart from one MPNST sample, which was an outlier, the tumours fell into two distinct groups (Figure 1A). The hierarchical clustering shows significantly differentially expressed miRNA separating MPNST and neurofibromas (Figure 1B). Sixteen miRNA were significantly differentially expressed in MPNST compared with neurofibromas and of these fourteen were downregulated in MPNST: these included miR-30e*, miR-29c*, miR-29c, miR-340*, miR-30c, miR-139-5p, miR-195, miR-151-5p, miR-342-5p, miR-146a, miR-150, miR-223, let-7 a and let-7 g with a false discovery rate of q=8.48E-03 for the least significant miRNA. In contrast, miR-210 and miR-339-3p were upregulated in MPNST compared with neurofibromas. The differential expression of five of these miRNA, including miR-139-5p, miR-210, miR-29c, miR-30e-3p, and miR-340 was confirmed by a TaqMan assay performed on the same source of RNA (extracted from frozen tissue sections) than for the microarray for the 19 index cases (the outlier was excluded) (Supplementary Figure S2A): the differential expression of let-7 g was not confirmed by the TaqMan assay.
MicroRNA profiling of neurofibromas and MPNST. (A) Multidimensional scaling of the 10 NF (light grey) and 9 MPNST (dark grey). (B) MicroRNA expression profiling and hierarchical clustering analysis of 10 neurofibromas and 9 MPNST representing the eight most significant differentially expressed miRNA with a cutoff of q=8.48E-03 for the least significant miRNA. Each row represents the relative levels of expression of a single miRNA and each column shows the expression levels for a single sample. (C) Heat-map of the miR-29 family members in 10 neurofibromas and 9 MPNST samples. All miR-29 family members were significantly differentially expressed with a P<0.05. The yellow and orange colours indicate relatively high and low expression, respectively.
We next validated our findings on a larger cohort of cases, 116 formalin-fixed paraffin-embedded PNSTs (56 neurofibromas and 60 MPNST), but this time focused on a small set of miRNAs, including miR-210, miR-29c, and miR-340, which are implicated in cancer (Supplementary Figure 2B). In view of miR-29c being a member of a larger family of miRNAs, we generated a supervised heat-map for miR-29 family including, miR-29a, b, c*, b-1*, and showed that these were significantly downregulated in MPNST compared with neurofibromas (Figure 1C). The star forms (*) of miRNA represent the opposite complementary strand of the guide strand: these are generally found to be less expressed compared with the non-star forms and are preferentially degraded, hence in future experiments we focus on the mature form (guide strand with no star) (Schwarz et al, 2003).
MiR-34a was not found to be significantly differentially expressed in MPNST when compared with neurofibromas as reported previously (Subramanian et al, 2010). Nevertheless, we found that 6 out of the 9 MPNST showed lower expression of miR-34a as compared with neurofibromas (Supplementary Figure S3).
miR-29 family targets extracellular matrix genes
Using prediction software/algorithm, TargetScanHuman (release 5.1) (http://www.targetscan.org/), PITA (http://genie.weizmann.ac.il/pubs/mir07/ mir07_data.html), PicTar (http://pictar.mdc-berlin.de/) and miRanda (http://www.microrna.org/microrna/home.do), available on the worldwide web, we identified a list of genes targeted by miR-29c and other family members. The target genes included extracellular matrix genes including collagen genes, MMPs, and the ADAM family of metalloprotease-disintegrins all of which are reported to be involved in cell migration and invasion (Supplementary Table S2 and S3) (Newby, 2006; Daley et al, 2008).
The full list of target genes present in both PITA and Targetscan predictions (Supplementary Table S3) were then subjected to pathway and gene function analysis using the GeneCodis2.0, a web server application integrating different sources of information such as Gene ontology (GO) and the Kyoto Encyclopaedia of Genes and Genomes (KEGG) databases, with the aim of finding modular patterns of interrelated annotations (Carmona-Saez et al, 2007; Nogales-Cadenas et al, 2009) (Supplementary Table S4). Analysis of the GO for biological process showed that extracellular matrix organisation was the most significantly represented with 13 genes, many of which are collagen genes, including COL2A1, COL5A1, COL4A6, COL11A1, COL5A2, with a corrected P-value of 4.09585E-09. The GO cellular component analysis also showed that collagen type IV and fibrillar collagens (type I, II, III, V, and XI) were significantly represented (P=5.68E-09 for GO:0005587 and P=7.10E-09 for GO:0005583, respectively) (Supplementary Table S4). Gene ontology terms for the molecular function, as well as the KEGG analysis, revealed that the predicted target genes were significantly enriched for genes involved in the metalloproteinase function including the ADAM family of metalloprotease-disintegrins including ADAMTS-9, 6, 17, 7, and 18, as well as MMP2 (P=0.00136053), and the ECM-receptor interaction (collagen genes) (P=7.14E-10) (Supplementary Table S4).
To validate the in silico predictions that ECM genes and metalloproteinase genes are targets of miR-29c in MPNST, the MPNST cell line sNF96.2 was transfected with miR-29c synthetic oligonucleotide (mimic). We chose sNF96.2 as a previous TaqMan assay showed that the sNF96.2 cell line had low miR-29c expression, a finding consistent with low miR-29c expression in MPNST. Analysis by RT–qPCR showed that the predicted genes including COL1A1, COL1A2, COL5A2, COL21A1, COL4A1, COL4A2, TGFβ3, and MMP2 were significantly downregulated compared with the scrambled miRNA mimic control (Figure 2A). These same genes were also differentially expressed in our previous GEM data generated from a set of NF and MPNST (Henderson et al, 2005) (Figure 2B).
miR-29c target genes. (A) sNF96.2 cells were transfected with either a miR-29c mimic or scrambled mimic in six-well plates. Two days post-transfection, cells were studied for expression of selected genes by RT–qPCR. Target genes expression is given as fold difference in sNF96.2 re-expressing miR-29c compared with the expression in the scrambled mimic cells. Bars represent mean and s.d. of three independent experiments relative to control (scrambled mimic). *P<0.01 and **P<0.001 to control. (B) Heat-map of selected target genes from our previously published GEM (Henderson et al, 2005). Expression levels are displayed in a colour spectrum that extends from light green (low expression) to bright red (high expression).
Functional effects of miR-29c on the sNF96.2 MPNST cell line
Transient transfection of the MPNST cell line, sNF96.2, using a miR-29c mimic, and scrambled oligonucleotides (control) revealed that cell motility was significantly reduced in transwell plates, and the scratch/wound healing assays (Figure 3).
miR-29c target genes involved in migration/invasion. sNF96.2 cells were transfected with either a miR-29c mimic, scrambled mimic, or oligofectamine only in six-well plates. (A, B) Two days post-transfection, cells were detached and ∼25 000 cells were transferred to individual wells of transwell plates. (A) Representative photomicrographs of cells transfected with miR-29c, scrambled mimic, and oligofectamine only on membranes. The cells were fixed and stained with gelRed after 6 h incubation. Migrated cells on the membrane were counted in four fields where the cells were evenly distributed. (B) The graph shows the average number of cells from each field (n=4) counted in three independent experiments (total n=12), the bars represent the mean. *P<0.05 miR-29c mimic vs control (scrambled mimic) and was calculated in GraphPad Prism doing paired t-test. (C) Representative images of the scratch assay 2 days post-transfection with miR-29c mimic and scrambled mimic. The images were monitored by time-lapse video-microscopy for ∼60 h. Photomicrographs were taken at time 0 (2 days post-transfection) and time 59:45 h.
No effect on cell growth/proliferation of the sNF96.2 cells was noted following transfection with the miR-29c mimics using a number of assays, including a live-cell imaging system IncuCyte, the MTS assay and growing the cells in 3-D culture using soft agar (Supplementary Figure S4).
Deficiency in proteolytic enzyme activity inhibits cell migration
As MMP2 has previously been shown to be a target of the miR-29 family (Steele et al, 2010), we speculated that the reduced migration in miR-29c-transfected sNF96.2 cells could be due, at least in part, to low levels of proteolytic enzyme activity. To test this we analysed the supernatant from miR-29c-transfected sNF96.2 cells by zymography. We found loss of bands corresponding to MMP2, which supports the hypothesis that the cells expressing miR-29c mimics had significantly reduced proteolytic activity compared with control cultures (Figure 4A). This interpretation was supported by the western blot and RT–qPCR analyses on the material from the same experiments, which showed a significant reduction in MMP2 protein level and mRNA, respectively, and also provides supportive evidence that MMP2 is a target gene of miR-29c (Figure 4B). In addition, we found that a heavier band was lost in the cells expressing miR-29c compared with the parental cell lines (Figure 4A). The molecular weight of this band corresponded to MMP9, which is not reported as a target of mir-29c. Western blot analysis and RT–qPCR of MMP9 showed no significant effect on the expression level of protein and mRNA, respectively. Furthermore, when the MPNST cell line sNF96.2 was transfected with miR-29c synthetic oligonucleotide (mimic), MMP9 expression was not altered (Figure 4B). These findings suggest that MMP9 is an indirect target of miR-29c. Finally, the zymography gels revealed that MMP3 activity was not affected when the cells were transfected with the miR-29c mimic (Figure 4A), a finding confirmed by western blot and RT–qPCR.
Proteolytic activity in sNF96.2 cells expressing miR-29c. (A) The supernatants from cells transfected with miR-29c and scrambled mimics were analysed by gelatin zymography in 0.1% gelatin–10% acrylamide gels with non-reducing conditions. Zones of gelatinolytic activity were revealed by negative staining. A different pattern of bands was observed when the cells were transfected with the miR-29c mimic and the scrambled mimic. A molecular weight ladder was used to identify, which MMPs was present or absent. Based on the molecular weight and the type of substrate (gelatin) used we concluded that the bands represented MMP9, MMP2, and MMP3 in the sNF96.2 cell line when transfected with the scrambled mimic, but that two of these corresponding to MMP2 and MMP9 were absent when cells were transfected with the miR-29c mimic. (B) Western blot analysis and RT–qPCR showed a reduction in MMP2 protein and mRNA expression in cells transfected with miR-29c mimic compared with control. In contrast MMP3 and MMP9 that are known not to be direct targets of miR-29c did not reveal a change in protein expression, and protein and mRNA expression, respectively.
Discussion
In this study we show that the majority of the miRNAs that are differentially expressed in MPNST compared with neurofibromas are downregulated in MPNST. We found it noteworthy that all family members of the miR-29 family, miR-29a, b, c*, b-1* are significantly downregulated, a finding which is consistent with their extensive sequence homology at the 5′-seed region, the sequence which is important for mRNA target recognition and specificity (Bartel, 2004). This is compatible with gene regulation being brought about by multiple miRNA exerting a synergistic effect (Doench and Sharp, 2004).
We focused specifically on miR-29c because it was more significantly differentially expressed compared with other miR-29 family members. The reproducibility of our findings in a larger cohort of 117 nerve sheath tumours, in addition to this miRNA being implicated in a number of other cancers including chronic lymphocytic leukaemia (Mraz et al, 2009; Stamatopoulos et al, 2009), oesophageal squamous cell carcinoma (Ding et al, 2011), nasopharyngeal carcinoma (Sengupta et al, 2008), lung cancer (Fabbri et al, 2007) made it an attractive molecule to study. Added to this was the availability of a MPNST cell line, sNF96.2 that we found to express low levels of miR-29c (Perrin et al, 2007) for which we confirmed the biallelic inactivation of neurofibromin 1.
The ECM is composed of a complex set of molecules that have an important role in the maintenance of tissue integrity. This is brought about by a finely tuned remodelling process achieved through breakdown and replacement of tissue and involves MMPs, which degrade various collagens, and non-collagenous ECM glycoproteins (Patterson et al, 2001). The ECM can act as a reservoir for growth factors/cytokines, which are involved in interactions between the ECM and components of the cell plasma membrane, for example, adhesion molecules and receptor tyrosine kinases. Hence, alteration of the constituent elements of ECM can result in modification of downstream transcriptional responses of various cytokines and bring about changes in proliferation, survival, and migration which account for the findings in disease (Russo et al, 2005; Newby, 2006; Daley et al, 2008). High levels of extracellular collagens and/or laminins have been shown to induce many tumour cell types to become more invasive in vitro, and to increase metastasis in animal models (Chintala et al, 1996; Song et al, 1997; Kuratomi et al, 1999; Menke et al, 2001; Kuratomi et al, 2002; Kaufman et al, 2005; Koenig et al, 2006; Shintani et al, 2006). Similarly, high levels of collagens and laminins have been associated with an increased likelihood of clinical metastasis in several types of human solid tumours (Ramaswamy et al, 2003). Consistent with this is that MMP2 overexpression has been shown to be involved in cell migration, and associated with the metastatic potential of malignant tumours (Egeblad and Werb, 2002). It is, therefore, of interest that the collagens types that are targeted by miR-29c are those that are also degraded by MMP2, and that both sets of genes are upregulated in MPNST as shown in our previously published GEM study (Henderson et al, 2005). These in silico data, together with our findings of reduced migration of MPNST cells in the presence of higher levels of miR-29c, argue that miR-29c acts as a tumour suppressor gene in MPNST and mediates it effect through modifying tumour migration by mediating its effect through both collagens and MMP2. It is also of interest that miR-29c was found to exert no effect on cell proliferation by a number of techniques, a result that is unsurprising as cell proliferation and migration are often mutually exclusive cellular phenomena (Zheng et al, 2009).
In search for a cause of the downregulation of miR-29c in MPNSTs, we reviewed the karyotype of the MPNST cell line, sNF96.2, (48, X, -X or Y, +7, add (7)(p22)x2, +8, add(9)(p24), +mar). However, these data do not suggest that there is loss of any of the miR-29 family members loci (on chromosomes 1 and 7) (Perrin et al, 2007). Moreover, our previous methylome analysis of the same two groups of samples (pooled) showed no significant differences in the miR-29c gene body (Feber et al, 2011), a result which was confirmed when the samples were analysed separately for gene body methylation by pyrosequencing (data not shown). Hence, there is no evidence to support that downregulation of miR-29c and other miR-29 family members in MPNST is caused by methylation.
Subramanian et al (2010) reported a significant downregultion of miR-34a in MPNST all of which exhibited a TP53 inactivation gene expression signature, compared with neurofibromas. They demonstrated that this was brought about by inactivation of TP53 using a cell line, a finding that is consistent with published data showing that miR-34a expression is regulated by TP53 (Chang et al, 2007; He et al, 2007). However, we were unable to reproduce this finding at a statistical level although we did shown that 6 out of the 9 MPNST showed lower expression of miR-34a, whereas lower expression was detected in only three neurofibromas. It is noteworthy that the TP53 gene expression signature of our 10 MPNSTs was unknown. In this context it is of interest that all of our patients had neurofibromatosis type 1, whereas it is not known if this were the case for the six patients in Subramanian’s study. This is relevant as TP53 mutations are reported to occur in ∼25% of MPNSTs, and that the minority of these (36%) are found in tumours from patients with neurofibromatosis type 1 (Verdijk et al, 2010). Hence, it is interesting to speculate that the different genetic background on which MPNSTs arise provide an explanation for the discrepant miR-34a results. An alternative explanation for the discrepancy between the two studies is that different miRNA microarray platform were used, and the small numbers of cases included in the studies could have compounded this. Nonetheless, it is reassuring that we reproduced the finding published by Subramanian et al (2010) that miR-29a was significantly downregulated in MPNST.
In summary, we have provided evidence that miR-29c has a pivotal role in the progression of nerve sheath tumours by increasing their invasive/migratory properties. Although the miRNA/RNA-I therapeutic field is in its infancy several approaches, including anti-miRNA oligonucleotides (Garzon et al, 2010), and antagomiRs – compounds that block the miRNA through complementarity (Garzon et al, 2010) and would restore miR-29c expression in MPNST represents a potential therapeutic approach for these tumours in the future.
Change history
05 March 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2): 281–297
Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6 (11): 857–866
Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A (2007) GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol 8 (1): R3
Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26 (5): 745–752
Chintala SK, Sawaya R, Gokaslan ZL, Rao JS (1996) The effect of type III collagen on migration and invasion of human glioblastoma cell lines in vitro. Cancer Lett 102 (1–2): 57–63
Colman SD, Williams CA, Wallace MR (1995) Benign neurofibromas in type 1 neurofibromatosis (NF1) show somatic deletions of the NF1 gene. Nat Genet 11 (1): 90–92
Daley WP, Peters SB, Larsen M (2008) Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci 121 (Part 3): 255–264
Di LG, Croce CM (2010) Roles of small RNAs in tumor formation. Trends Mol Med 16 (6): 257–267
Ding DP, Chen ZL, Zhao XH, Wang JW, Sun J, Wang Z, Tan FW, Tan XG, Li BZ, Zhou F, Shao K, Li N, Qiu B, He J (2011) miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis 32 (7): 1025–1032
Doench JG, Sharp PA (2004) Specificity of microRNA target selection in translational repression. Genes Dev 18 (5): 504–511
Egeblad M, Werb Z (2002) New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2 (3): 161–174
Esquela-Kerscher A, Slack FJ (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6 (4): 259–269
Evans DG, Baser ME, McGaughran J, Sharif S, Howard E, Moran A (2002) Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet 39 (5): 311–314
Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM (2007) MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA 104 (40): 15805–15810
Feber A, Wilson GA, Zhang L, Presneau N, Idowu B, Down TA, Rakyan VK, Noon LA, Lloyd AC, Stupka E, Schiza V, Teschendorff AE, Schroth GP, Flanagan A, Beck S (2011) Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res 21 (4): 515–524
Fletcher DM, Unni KK, Mertens F (2002) World Health Organization Classification of Tumours Pathology and Genetics of Tumours of Soft Tissue and Bone. International Agency for Research on Cancer (IARC): Lyon
Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM (2006) MicroRNA expression and function in cancer. Trends Mol Med 12 (12): 580–587
Garzon R, Marcucci G, Croce CM (2010) Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 9 (10): 775–789
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 (10): R80
Gutmann DH, Aylsworth A, Carey JC, Korf B, Marks J, Pyeritz RE, Rubenstein A, Viskochil D (1997) The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA 278 (1): 51–57
He L, He X, Lim LP, de SE, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ (2007) A microRNA component of the p53 tumour suppressor network. Nature 447 (7148): 1130–1134
Henderson SR, Guiliano D, Presneau N, McLean S, Frow R, Vujovic S, Anderson J, Sebire N, Whelan J, Athanasou N, Flanagan AM, Boshoff C (2005) A molecular map of mesenchymal tumors. Genome Biol 6 (9): R76
Karube K, Nabeshima K, Ishiguro M, Harada M, Iwasaki H (2006) cDNA microarray analysis of cancer associated gene expression profiles in malignant peripheral nerve sheath tumours. J Clin Pathol 59 (2): 160–165
Kaufman LJ, Brangwynne CP, Kasza KE, Filippidi E, Gordon VD, Deisboeck TS, Weitz DA (2005) Glioma expansion in collagen I matrices: analyzing collagen concentration-dependent growth and motility patterns. Biophys J 89 (1): 635–650
King AA, Debaun MR, Riccardi VM, Gutmann DH (2000) Malignant peripheral nerve sheath tumors in neurofibromatosis 1. Am J Med Genet 93 (5): 388–392
Kleihues P, Cavenee WK (2000) World Health Organization Classification of Tumours Pathology and Genetics of Tumours of Nervous System. International Agency for Research on Cancer (IARC): Lyon
Koenig A, Mueller C, Hasel C, Adler G, Menke A (2006) Collagen type I induces disruption of E-cadherin-mediated cell-cell contacts and promotes proliferation of pancreatic carcinoma cells. Cancer Res 66 (9): 4662–4671
Kroep JR, Ouali M, Gelderblom H, Le CA, Dekker TJ, Van GM, Hogendoorn PC, Hohenberger P (2011) First-line chemotherapy for malignant peripheral nerve sheath tumor (MPNST) versus other histological soft tissue sarcoma subtypes and as a prognostic factor for MPNST: an EORTC soft tissue and bone sarcoma group study. Ann Oncol 22 (1): 207–214
Kuratomi Y, Nomizu M, Nielsen PK, Tanaka K, Song SY, Kleinman HK, Yamada Y (1999) Identification of metastasis-promoting sequences in the mouse laminin alpha 1 chain. Exp Cell Res 249 (2): 386–395
Kuratomi Y, Nomizu M, Tanaka K, Ponce ML, Komiyama S, Kleinman HK, Yamada Y (2002) Laminin gamma 1 chain peptide, C-16 (KAFDITYVRLKF), promotes migration, MMP-9 secretion, and pulmonary metastasis of B16-F10 mouse melanoma cells. Br J Cancer 86 (7): 1169–1173
Legius E, Marchuk DA, Collins FS, Glover TW (1993) Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat Genet 3 (2): 122–126
Menke A, Philippi C, Vogelmann R, Seidel B, Lutz MP, Adler G, Wedlich D (2001) Down-regulation of E-cadherin gene expression by collagen type I and type III in pancreatic cancer cell lines. Cancer Res 61 (8): 3508–3517
Miller SJ, Rangwala F, Williams J, Ackerman P, Kong S, Jegga AG, Kaiser S, Aronow BJ, Frahm S, Kluwe L, Mautner V, Upadhyaya M, Muir D, Wallace M, Hagen J, Quelle DE, Watson MA, Perry A, Gutmann DH, Ratner N (2006) Large-scale molecular comparison of human schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res 66 (5): 2584–2591
Mraz M, Malinova K, Kotaskova J, Pavlova S, Tichy B, Malcikova J, Stano KK, Smardova J, Brychtova Y, Doubek M, Trbusek M, Mayer J, Pospisilova S (2009) miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia 23 (6): 1159–1163
Newby AC (2006) Matrix metalloproteinases regulate migration, proliferation, and death of vascular smooth muscle cells by degrading matrix and non-matrix substrates. Cardiovasc Res 69 (3): 614–624
Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A (2009) GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res 37, Web Server issue): W317–W322
Pasquinelli AE, Hunter S, Bracht J (2005) MicroRNAs: a developing story. Curr Opin Genet Dev 15 (2): 200–205
Patterson ML, Atkinson SJ, Knauper V, Murphy G (2001) Specific collagenolysis by gelatinase A, MMP-2, is determined by the hemopexin domain and not the fibronectin-like domain. FEBS Lett 503 (2–3): 158–162
Perrin GQ, Li H, Fishbein L, Thomson SA, Hwang MS, Scarborough MT, Yachnis AT, Wallace MR, Mareci TH, Muir D (2007) An orthotopic xenograft model of intraneural NF1 MPNST suggests a potential association between steroid hormones and tumor cell proliferation. Lab Invest 87 (11): 1092–1102
Porter DE, Prasad V, Foster L, Dall GF, Birch R, Grimer RJ (2009) Survival in malignant peripheral nerve sheath tumours: a comparison between sporadic and neurofibromatosis type 1-associated tumours. Sarcoma 2009: 756395
Presneau N, Shalaby A, Idowu B, Gikas P, Cannon SR, Gout I, Diss T, Tirabosco R, Flanagan AM (2009) Potential therapeutic targets for chordoma: PI3K/AKT/TSC1/TSC2/mTOR pathway. Br J Cancer 100 (9): 1406–1414
Ramaswamy S, Ross KN, Lander ES, Golub TR (2003) A molecular signature of metastasis in primary solid tumors. Nat Genet 33 (1): 49–54
Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23 (20): 2700–2707
Russo VC, Schutt BS, Andaloro E, Ymer SI, Hoeflich A, Ranke MB, Bach LA, Werther GA (2005) Insulin-like growth factor binding protein-2 binding to extracellular matrix plays a critical role in neuroblastoma cell proliferation, migration, and invasion. Endocrinology 146 (10): 4445–4455
Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115 (2): 199–208
Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P (2008) MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci USA 105 (15): 5874–5878
Shintani Y, Hollingsworth MA, Wheelock MJ, Johnson KR (2006) Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH(2)-terminal kinase 1 and up-regulating N-cadherin expression. Cancer Res 66 (24): 11745–11753
Song SY, Nomizu M, Yamada Y, Kleinman HK (1997) Liver metastasis formation by laminin-1 peptide (LQVQLSIR)-adhesion selected B16-F10 melanoma cells. Int J Cancer 71 (3): 436–441
Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L (2009) microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood 113 (21): 5237–5245
Steele R, Mott JL, Ray RB (2010) MBP-1 upregulates miR-29b that represses Mcl-1, collagens, and matrix-metalloproteinase-2 in prostate cancer cells. Genes Cancer 1 (4): 381–387
Subramanian S, Thayanithy V, West RB, Lee CH, Beck AH, Zhu S, Downs-Kelly E, Montgomery K, Goldblum JR, Hogendoorn PC, Corless CL, Oliveira AM, Dry SM, Nielsen TO, Rubin BP, Fletcher JA, Fletcher CD, van de Rijn M (2010) Genome-wide transcriptome analyses reveal p53 inactivation mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours. J Pathol 220 (1): 58–70
Verdijk RM, den Bakker MA, Dubbink HJ, Hop WC, Dinjens WN, Kros JM (2010) TP53 mutation analysis of malignant peripheral nerve sheath tumors. J Neuropathol Exp Neurol 69 (1): 16–26
Zheng PP, Severijnen LA, van der Weiden M, Willemsen R, Kros JM (2009) Cell proliferation and migration are mutually exclusive cellular phenomena in vivo: implications for cancer therapeutic strategies. Cell Cycle 8 (6): 950–951
Acknowledgements
We are grateful to the individuals who participated in the research and to the clinicians, and support staff in the London Sarcoma Service involved in their care. We are also grateful to Dr Eve Coulter and Dr Bernadine Idowu for helping in processing the miRNA microarrays. The research was funded by Skeletal Cancer Action Trust (SCAT) UK. This research benefitted from the Royal National Orthopaedic Hospital Musculoskeletal Research Programme and Biobank, and the University College London Hospital and University College London Comprehensive Biomedical Research Programme.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies the paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Presneau, N., Eskandarpour, M., Shemais, T. et al. MicroRNA profiling of peripheral nerve sheath tumours identifies miR-29c as a tumour suppressor gene involved in tumour progression. Br J Cancer 108, 964–972 (2013). https://doi.org/10.1038/bjc.2012.518
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2012.518
Keywords
This article is cited by
-
MicroRNA-155 contributes to plexiform neurofibroma growth downstream of MEK
Oncogene (2021)
-
Deregulated microRNAs in neurofibromatosis type 1 derived malignant peripheral nerve sheath tumors
Scientific Reports (2020)
-
Tumor miRNA expression profile is related to vestibular schwannoma growth rate
Acta Neurochirurgica (2020)
-
Hepatitis B virus promotes cancer cell migration by downregulating miR-340-5p expression to induce STAT3 overexpression
Cell & Bioscience (2017)
-
MiR-216b inhibits cell proliferation by targeting FOXM1 in cervical cancer cells and is associated with better prognosis
BMC Cancer (2017)