Abstract
Aberrant chromatin remodeling is involved in the pathogenesis of Huntington's disease (HD) but the mechanism is not known. Herein, we report that mutant huntingtin (mtHtt) induces the transcription of alpha thalassemia/mental retardation X linked (ATRX), an ATPase/helicase and SWI/SNF-like chromatin remodeling protein via Cdx-2 activation. ATRX expression was elevated in both a cell line model and transgenic model of HD, and Cdx-2 occupancy of the ATRX promoter was increased in HD. Induction of ATRX expanded the size of promyelocytic leukemia nuclear body (PML-NB) and increased trimethylation of H3K9 (H3K9me3) and condensation of pericentromeric heterochromatin, while knockdown of ATRX decreased PML-NB and H3K9me3 levels. Knockdown of ATRX/dXNP improved the hatch rate of fly embryos expressing mtHtt (Q127). ATRX/dXNP overexpression exacerbated eye degeneration of eye-specific mtHtt (Q127) expressing flies. Our findings suggest that transcriptional alteration of ATRX by mtHtt is involved in pericentromeric heterochromatin condensation and contributes to the pathogenesis of HD.
Similar content being viewed by others
Main
Huntington's disease (HD) is an incurable and fatal autosomal dominant neurodegenerative disorder of mid-life onset characterized by chorea and progressive cognitive decline.1 HD is caused by expanded CAG repeats coding for glutamine (Q) in the exon 1 of the Huntingtin (Htt) gene.2 One mechanism by which mutant Htt (mtHtt) contributes to neurodegeneration is through transcriptional dysregulation and chromatin remodeling.3, 4, 5, 6 Aberrant nucleosomal dynamics have recently been implicated in the pathogenesis of HD but the exact role of chromatin remodeling remains to be elucidated.
Chromatin remodeling complexes modulate the structure of nucleosomes around the transcription initiation site and define its accessibility to the transcription machinery in an adenosine triphosphate (ATP) -dependent manner.7 Alpha thalassemia/mental retardation X linked (ATRX) is a DNA-dependent ATPase/helicase belonging to the Rad54-like subfamily of SWI/SNF chromatin remodeling proteins.8, 9 Mutations of ATRX are directly linked to ATR-X syndrome, a rare X-linked mental retardation disorder accompanied with learning difficulties, characteristic facial appearance, anemia, and genomic instability.10, 11 Transgenic expression of ATRX results in growth retardation and neural tube defects, suggesting that normal development of the cortex are susceptible to ATRX dosage.12 ATRX collaborates with methyl-CpG-binding protein 2 and Daxx and disruption of these interactions may cause transcriptional deregulation and pathological changes that contribute to mental retardation.13, 14, 15 In addition, the loss of ATRX leads to genomic instability and impaired corticogenesis.11, 16 ATRX is an epigenetic modulator necessary for cell survival during early neuronal differentiation and its abnormal induction or deficiency leads to neuronal degeneration, but the mechanisms responsible are largely unknown.
Given that deregulation of nucleosomal dynamics contributes to the HD pathogenesis and that ATRX expression is elevated in peripheral blood of HD, it is reasonable to suggest that ATRX could have a role in chromatin remodeling and neurodegeneration in HD. Accordingly, in order to determine how ATRX induction is linked to chromatin remodeling and the neurodegenerative process, we investigated transcriptional mechanisms of ATRX gene expression in HD. As ATRX null mice do not survive beyond birth, we crossed mtHtt (Q127) and ATRX/dXNP transgenic flies to examine the effects of ATRX depletion on embryonic development and eye phenotype in the context of triplet repeat expansion.
Results
ATRX is elevated in a cell line and an animal model of HD
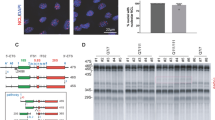
In order to investigate whether ATRX is linked to histone remodeling in the pathogenesis of HD, we first examined the protein and the immunoreactivity of ATRX in a striatal cell line model and an animal (R6/2) of HD17 (Figure 1 and Supplementary Figure 1). The immortalized striatal cells were previously established from STHdhQ7/Q7 (wild type) and homozygous mutant STHdhQ111/Q111 (Q111/Q111) knock-in mouse embryos.18 The level of ATRX protein was significantly increased in STHdhQ111/Q111 compared with STHdhQ7/Q7 (Q7/Q7) control cells, as shown by western blot analysis (Figure 1a). Consistent with western blot data, ATRX immunoreactivity was elevated, and punctate ATRX positive structures were markedly increased in the nucleus of Q111/Q111 cells (Figure 1b). ATRX immunoreactivity was also elevated in a mouse model of HD (R6/2 line). The R6/2 line is a transgenic mouse line expressing exon 1 of the Htt gene with an expanded CAG repeat (150 CAGs). The mice develop similar clinical features, along with striatal atrophy and neuronal intranuclear inclusions that are similar to HD in humans. (Figure 1c and Supplementary Figure 1a). We next determined the distribution of ATRX immunoreactivity in striatal neurons R6/2 mice using confocal microscopy and image analysis. We found that ATRX immunoreactivity was increased, and that the majority of ATRX immunoreactivity was colocalized with trimethylation of H3K9 (H3K9me3) in prominent clusters of pericentromeric heterochromatin (chromocenters) in R6/2 mice (Figure 1c). ATRX immunoreactivity also colocalized with HP1α in nuclear foci of R6/2 mice that were spatially different and distinguishable from WT mice (Figure 1d and Supplementary Figure 1b). To determine whether ATRX was affected in vivo, we examined ATRX mRNA and protein levels in striatal neurons stably expressing GFP or the mutant huntingtin (mtHtt) GFP-fusion protein (GFP-Q79) and found that ATRX levels were increased in mutant cells compared with the control cells (Figures 1e and f).
ATRX is altered in HD (STHdhQ111/111) cells and HD (R6/2) animals. (a) ATRX protein is markedly increased in STHdhQ111/111 (Q111/Q111) cells (upper panel). Densitometric analysis of ATRX protein from four separate experiments (bottom panel) (*significant at P<0.007). ATRX was normalized to β-actin. (b) The size of ATRX foci is enlarged in the nucleus of Q111 cells in comparison to STHdhQ7/Q7 (Q7/Q7) cells (*significant at P<0.001). (c) ATRX is increased in an animal model of HD (R6/2 line). Deconvolved images (upper panels) show intense staining of heterochromatin condensation by ATRX (green) and H3K9me3 (red) in striatal neuron of HD (R6/2) mice compared with wild-type mice. Isosurface images (middle panels) illustrate clear spatial patterns for the expansion of chromocenters and the condensation of ATRX (green) and H3K9me3 (red)-positive heterochromatin structures. The intensity of ATRX and H3K9me3 is increased in R6/2 mice compared with WT (bottom panel). Arrows (white) indicate the spots of line measurement. Scale bar: 10 μm. (d) Colocalization of ATRX and hetrochromatin-binding protein α (HP1α) is increased in striatal neurons of HD (R6/2) mice. Scale bar: 10 μm. (e) mtHtt (GFP-Q79) increases the protein level of ATRX. (f) mtHtt (GFP-Q79) induces the expression of ATRX gene in Q7 striatlal cells (*significant at P<0.02). ATRX mRNA was normalized to GAPDH mRNA. Data are average±S.E. of six experiments
Cdx2 drives ATRX promoter activity
To determine how ATRX is activated at the transcriptional level, we cloned the promoter of human ATRX into a luciferase vector (pGL4.14). We ran TFSEARCH software (Kyoto University, Japan) with the promoter sequence of ATRX and found transcription factor-binding elements for Cdx, Sp1/3, CREB, and GATA2-binding elements (Figure 2a). To determine which transcription factors modulate the transcriptional activation of ATRX, we constructed variants of the ATRX promoter region (ATRX, −548/+101; ATRX, −438/+101; ATRX, −248/+101). We co-transfected Sp1, Sp3, Cdx1, and Cdx2 expression vectors with the ATRX promoter plasmids and measured luciferase activity. Among four factors, Cdx2 showed the strongest transcriptional activity of the ATRX promoter (Figure 2b). It is well known that Cdx1 and Cdx2 transcription factors are important regulators of intestinal epithelial cell differentiation, apoptosis, and proliferation but its function in the central nervous system has not been investigated. While two potential Cdx-binding sites emerged, our data demonstrates that the proximal site functions as an important cis-element within the ATRX promoter (Figures 2c and d). Moreover, Cdx2 induced the transcription of ATRX in a dose-dependent manner more potently than Cdx1 (Figure 2e). As we identified that human and mouse promoter of ATRX contain Cdx consensus-binding site (Supplementary Figure 2a), we performed chromatin immunoprecipitation (ChIP) assays to detect if Cdx2-DNA occupancy was altered within the ATRX gene promoter in HD (R6/2) mice. The Cdx2-DNA occupancy was increased approximately ninefold in the neostriatal tissues of HD (R6/2) mice compared with the controls (Figure 2f). In contrast, Sp1- and Sp3-DNA occupancy was slightly but not significantly increased in the HD (R6/2) mice (Supplementary Figures 2b and c). Taken together, ChIP and luciferase assays indicate that Cdx2 is a key transcriptional regulator of the ATRX gene in vivo. This promoter assay data provides further evidence that Cdx2 is an important transcription factor affecting the transcription of the ATRX gene. Protein levels of Cdx2 were increased in striatal neurons of HD (R6/2) mice compared with WT mice by western blot (Figure 2g). Immunofluorescence staining and confocal microscopy showed that Cdx2 immunoreactivity is elevated in the nucleus of striatal neurons in R6/2 mice (Figure 2h). As we expected, overexpression of Cdx2 increased the expression of ATRX gene as monitored by quantitative real time RT-PCR (Figure 2i). On the other hand, knockdown of Cdx2 with shRNA reduced the levels of ATRX, Cdx2, and H3K9me3 (Figure 2j, Supplementary Table 1).
ATRX is induced by mtHtt through Cdx2 activation. (a) The DNA sequence of human ATRX promoter shows putative transcription factor binding elements. (b) Cdx1, Cdx2, Sp1, and Sp3 induce the transcriptional activity of ATRX promoter. SH-SY5Y cells were transiently co-transfected with of ATRX promoter (−548/+101) (400 ng/ml) and Cdx1, Cdx2, Sp1, and Sp3 (200 ng/ml) plasmids. (c) Nested deletion constructs of human ATRX 5′-UTR promoter. ATRX promoter activity was determined using a series of deletion reporter constructs (ATRX-548/+101; ATRX-438/+101; ATRX-248/+101). (d) Proximal Cdx-binding site is essential for the basal and Cdx2-induced ATRX promoter activity. All luciferase data represent the mean±S.E. of three independent experiments. (e) Cdx2 activates ATRX promoter in a dose-dependent manner. (f) Cdx2-DNA occupancy is increased in striatal neurons of HD (R6/2) mice (n=3) in comparison to control (n=3). **Significant at P<0.01. (g) Cdx2 and ATRX levels are elevated in striatal neurons of HD (R6/2) mice (n=3) in comparison to control (n=3). **Significant at P<0.01. (h) Cdx2 immunoreactivity is increased in the nucleus of HD (R6/2) striatal neurons. (i) Cdx2 increases the level of ATRX mRNA in SH-SY5Y cells. Data are average ±S.E. of three experiments. **Significant at P<0.01. (j) Knockdown of Cdx2 reduces the level of ATRX protein and H3K9me3. Data are the representative of two experiments
Consistent with our earlier finding that mtHtt could increase Atrx protein levels, ATRX promoter activity was similarly increased in cells expressing mtHtt (Q103) compared with the control cell lines (GFP and GFP-27) (Figure 3a). To verify the role of Cdx2 as a transcriptional activator over the proximal region of Cdx-binding site in the ATRX promoter, we constructed a deletion mutant for the predicted Cdx-binding site using site-directed mutagenesis (Figure 3b). As a template plasmid, the ATRX (−438/+101) promoter variant was used. The mutant ATRX promoter (Cdx-binding element deletion) showed a basal promoter activity close to plasmid (pGL4.14) control levels (Figure 3b). As we expected, the deletion of Cdx2-binding site also abrogated the mtHtt-induced promoter activity of the ATRX gene (Figure 3c). Thus, our data indicates for the first time that Cdx2 is activated by mtHtt in the central nervous system and that it regulates mtHtt-mediated ATRX transcription.
mtHtt induces ATRX promoter activity. (a) mtHtt (GFP-Q103) increased ATRX-promoter activity but not GFP-Q27 in SH-SY5Y cells. *Significantly increased in comparison to GFP control at P<0.01. (b) Mutation of Cdx2-binding site abolishes Cdx2-dependent activation of ATRX transcription. **Significantly decreased in comparison to the control at P<0.001. (c) The deletion of Cdx-binding element abolishes mtHtt (Q79)-induced ATRX promoter activity. *Significantly decreased in comparison to wild-type ATRX promoter vector (−438/+101) at P<0.01. All luciferase data represent the mean±S.E. of three independent experiments
ATRX modulates promyelocytic leukemia-nuclear bodies (PML-NBs) and heterochromatin condensation
ATRX is localized to pericentromeric heterochromatin and can physically interact with proteins that reside within heterochromatin including HP1.19 The ATRX protein also localizes to PML-NBs that may have a role in transcription regulation during the neurodegenerative process.20 To address how ATRX contributes to PML-NB formation and chromatin remodeling, we overexpressed GFP-ATRX fusion protein in striatal cells and measured PML-NB and H3K9me3 immunoreactivities and levels. PML-positive small nuclear speckles were present in Q7/Q7 cells (Figure 4a). Overexpression of ATRX increased the size of PML-NBs (Figures 4a and b). In contrast, knockdown of ATRX by siRNA reduced the number of PML-NBs (Figure 4c). Overexpression of ATRX was also associated with increased H3K9me3 immunoreactivity within the nuclei of Q7/Q7 cells (Figure 4d) that colocalized with ATRX (Figure 4d). Both the N-terminal and C-terminal domains of ATRX were involved in the hypertrimethylation of H3K9 and heterochromatin condensation in human neuroblastoma cells (Supplementary Figure 3a). Interestingly, overexpression of the C-terminal helicase domain of ATRX, including NLS (GFP-ATRX NLS1204-2492), resulted in abnormal heterochromatin condensation and nuclear damage in Q7/Q7 and Q111/Q111 striatal cells (Supplementary Figure 3b). In contrast, knockdown of ATRX reduced the H3K9me3 foci and pericentromeric nuclear condensation in Q111/111 and Q7/Q7 cells (Supplementary Figure 4). Western analysis showed that ATRX overexpression increases H3K9me3 levels in a dose-dependent manner (Figure 4e). Conversely, knockdown of ATRX by siRNA reduced the level of H3K9me3 in a dose-dependent manner (Figure 4f).
ATRX modulates PML nuclear body (PML-NB) and pericentromeric heterochromatin condensation. (a) Overexpression of ATRX (green) increases the PML-NB immunoreactivity (red) in Q7/Q7 striatal cells. (b) ATRX expands the size of PML-NBs. GFP-positive cells (n=48), GFP-ATRX-positive cells (n=38). Quantitative image analysis showed a significant increase in the size of PML-NBs. *Significant at P<0.01. (c) Knockdown of ATRX with siRNA reduces the level of PML-NBs (red). (d) ATRX (green) increases H3K9me3 immunoreactivity (red), a marker of pericentromeric heterochromatin condensation. (e) ATRX elevates the level of H3K9me3 in a does-dependent manner. (f) Knockdown of ATRX with siRNA reduces the level of ATRX and H3K9me3 in comparison to siRNA control
ATRX/dXNP is implicated in mtHtt-mediated toxicity
ATRX knockout mice are lethal and CNS-restricted knockout animals die at birth making it difficult to study the role of ATRX in HD in this model. To further define the role of ATRX in HD, we used a Drosophila model of mtHtt-mediated toxicity (Figure 5). It has previously been shown that mtHtt-induced neurodegeneration is mimicked in Drosophila with a GAL4/UAS system and a transgene expressing 127 consecutive glutamines (127Q).21 The genetic cross with a line that expresses GAL4/UAS-poly127Q under the control of the elav promoter led to a significant (35%) reduction of hatching rate (Figure 5a). ATRX/dXNP knockdown (XNPC1/+) improved the hatching rate affected by mtHtt (127Q). mtHtt (127Q) expression markedly deformed the fly embryo and ATRX/dXNP knockdown (XNPC1/+) significantly reduced the number of deformed embryos (Figures 5b and c). We also established a line that expresses GAL4 under the control of the glass multimer repeat promoter that resulted in the expression of 127Q in all cells of the retina and led to eye-specific degeneration (Figure 5d). To further investigate the role of ATRX/dXNP in mtHtt-mediated toxicity, we crossed the GMRGAL4/UAS-127Q flies with line EP635, which carries a P(EP)-element insertion +20 bp from transcription site of ATRX/dXNP. The P(EP) insertion resulted in increased ATRX/dXNP expression in the presence of GAL4. As shown in Figure 5d, external eye pigmentation and the regular array of ommatidia were disrupted by mtHtt (127Q). The double expression of ATRX/dXNP exacerbated mtHtt (127Q)-mediated degeneration with complete loss of eye pigmentation.22, 23 Flies that only overexpressed ATRX/dXNP resulted in no significant loss of eye pigmentation and minor disruption in the regular array of ommatidia (Figure 5d).
ATRX/dXNP modulates mtHtt (127Q)-induced pathology in a fly model. (a) ATRX/dXNP knockdown rescues the hatching rate of fly embryos with mtHtt (127Q) overexpression. *Significantly different at P <0.001 (n≥8). (b) ATRX/dXNP knockdown improves embryonic development affected by mtHtt (127Q). Fly embryos (stage 14) were stained with DAPI. (c) ATRX/dXNP knock-down restores embryonic development affected by mtHtt (127Q). *Significantly different at P<0.001 (n≥4). (d) ATRX/dXNP overexpression exacerbates mtHtt (127Q)-induced eye degeneration. Control fly expressed the transcription factor GAL4 under control of the eye-specific promoter GMR. Expression of mtHtt (127Q) leads to progressive eye degeneration. Upregulation of ATRX/dXNP expression in concert with 127Q exacerbated the disruption of ommatidium and eye degeneration in comparison to control. Genotypes: CTL (elav-GAL4), p127Q (elav-GAL4/UAS-poly127Q), XNPC1 (elav-GAL4/+;XNPC1/+), p127Q XNPC1(elav-GAL4/ UAS-poly127Q; XNPC1/+), w1118 (w1118/w1118), GMR>p127Q (GMR-GAL4/UAS-poly127Q), GMR>p127Q/+; XNPep/+ (GMR-GAL4/UAS-poly127Q; XNPep/+), GMR-GAL4/+; XNPep/+ (GMR-GAL4/+; XNPep/+)
Discussion
ATRX is known to be crucial for normal development and organization of the central nervous system. Overexpression of ATRX in transgenic mice leads to craniofacial anomalies, seizures, and behavioral changes.12 It has been reported that ATRX expression levels are elevated in the white blood cells of presymptomatic (1.6-fold) and symptomatic HD (2-fold) patients compared with the controls.24 Given that deregulation of nucleosomal dynamics contributes to the HD pathogenesis and that ATRX expression is elevated in the peripheral blood of HD, it is reasonable to suggest that ATRX could have a role in chromatin remodeling and neurodegeneration in HD. Indeed, we found that ATRX is transcriptionally induced by mtHtt through activation of Cdx2. Our ChIP assays confirmed that the preferential binding of Cdx2 to the ATRX promoter is markedly increased in HD (R6/2) and that Cdx2 overexpression directly regulates the expression of ATRX. Increased ATRX expression in turn leads to PML-NB expansion and increased H3K9me3.20 In contrast, Cdx2 knockdown using shRNA leads to reduced ATRX and H3K9me3 levels. Cdx2 knockdown also abrogates the mtHtt-induced promoter activity of the ATRX gene. Our data indicate that activation of Cdx2 by mtHtt is closely linked to expression of the ATRX gene. DNA occupancy by both Sp1 and Sp3 in the promoter region indicate that Sp3 might also be involved in the transcriptional activation of ATRX in HD. Given the presence of a CREB-binding site in the proximal 5′-UTR promoter region of the ATRX gene, it is likely that CREB may also participate in the transcriptional regulation of ATRX.
Knockdown of ATRX reduced the formation of PML-NBs and decreased heterochromatin condensation by reducing H3K9me3 levels in striatal cells. These results suggest that heterochromatin remodeling in striatal cells is driven by ATRX under neurodegenerative conditions.19 Interestingly, both the N-terminal ATRX-Dnmt3-Dnmt3L (ADD) domain and the C-terminal ATPase/helicase domain of ATRX modulated the dynamics of pericentromeric heterochromatin by increasing H3K9me3 levels at its target loci and ultimately, to nuclear damage in striatal cells.15 These findings suggest that ATRX facilitates the pathological heterochromatin condensation and triggers the cascade of events leading to neuronal degeneration.12 A recent study confirms that the ADD domain of ATRX interacts with histone H3 tails that are trimethylated at lysine 9.25
To further examine whether ATRX directly contributes to the pathological process in HD, we developed a Drosophila model of ATRX/dXNP and characterized its physiological effects by crossing ATRX/dXNP and mtHtt (127Q) flies. We found that knockdown of ATRX increased the hatching rate and reduced egg deformations caused by mtHtt (127Q). Moreover, overexpression of ATRX/dXNP exacerbated eye degeneration in the HD fly.22, 23 In contrast, our results show that, in part, the deformations within the fly egg and the degeneration of fly eyes are associated with mtHtt, and the pathological phenomena are mediated by gain of function of ATRX. Despite our demonstration of linking ATRX to the pathogenesis of HD via the formation of heterochromatin condensation and PML-NBs formation, the identification of ATRX targets in HD remains to be determined with a high priority.
Taken together, the present study validates the contribution of ATRX to HD pathogenesis through epigenetic modulation of heterochromatin condensation (Figure 6). ATRX participates in organizing the structure of pericentromeric heterochromatin by elevating trimethylated histone H3K9 levels in striatal cells. Our findings suggests that ATRX-mediated epigenetic modifications contribute significantly to pathogenesis of neurodegeneration in HD and that neuronal ATRX levels may be an indicator of aberrant chromatin remodeling in neurological disorders.
Schematic diagram illustrating the potential role of mtHtt as an activator of the Cdx2 transcriptional factor and, in turn, increased ATRX transcription and heterochromatin condensation. Increased ATRX expression causes H3K9 hyper-trimethylation and pericentromeric chromatin condensation. These aberrant epigenetic changes affect the function of striatal neurons and may contribute to the pathogenesis of HD
Materials and Methods
Plasmids and cell culture
pcDNA3/GFP-ATRX (1-745), ATRX (1201-2491), and ATRX (NLS1916-2492) were generously provided from Dr. Bird,15 and pcDNA3-Cdx1 and Cdx2 constructs were from Dr. Iovanna.26 STHdhQ7/7 (wild type) and STHdhQ111/111 (HD knock-in striatal cell line expresses mtHtt at endogenous level), were generously provided from Dr. MacDonald.18 SH-SY5Y were maintained and used for ATRX promoter assay.
Fly (Drosophila) model of HD (127Q) and ATRX/dXNP
Flies were maintained on a standard mixture of corn meal, yeast, and agar at room temperature. The UAS-127Q and GMR-GAL4 flies were described previously.27 The EP635 flies were also described previously.22, 23 By imprecise excision of a P-element in the EP635 line, we generated XNPC1, which has deletion of 1065 bp, removing the first exon, including the translational start site of ATRX/XNP.
Analysis of Drosophila development
Fifty embryos of each genotype were placed on grape juice agar plates. After incubation for 2 days at 25 °C, the number of hatched larvae was counted to determine embryonic lethality. The experiment was repeated eight times (n=400), and statistically analyzed by a Student's t-test. To visualize developing embryos, Hoechst 33 258 staining was performed. Briefly, embryos were collected for 16 h at 25 °C, and dechorionated in 50% bleach for 3 min. The dechorionated embryos were fixed in 4% paraformaldehyde with heptane for 20 min and devitellinized with methanol. After several washes with PBT (PBS with 0.1% Tween 20), the embryos were incubated in PBT with Hoechst 33 258 for 5 min at 25 °C. The samples were examined under an Axiophot2 fluorescent microscope (Carl Zeiss AG, Jena, Germany).
Animals
Male R6/2 mice were bred with females from their background strain (B6CBAFI/J), and offspring were genotyped using PCR.5, 17 CAG repeat length remained stable within a 147–153 range. Female mice were used in the experimental paradigms.
Quantitative real time-PCR (qRT-PCR)
qRT-PCR was performed as previously described.5 The primer sequences for ATRX gene expression as follow: forward, 5′-CAGAGAAATGTAGACCCAGA-3′; reverse, 5′-GGTTATCAGTAGCACAGTCA-3′. The primer sequences for ChIP-qPCR of ATRX promoter was as follow: forward, 5′-AGGATCTGTGTAGTGGTCGA-3′; reverse, 5′-CTGCTGACCTCAAAGTGTCA-3′.
Chromatin immunoprecipitation (ChIP)
ChIP for Cdx2, Sp1, and Sp3 binding to DNA was performed using a ChIP assay kit (Santa Cruz Biotech, Santa Cruz, CA, USA) as previously described.28 Tissues and cells were crossed-linked with 1% formaldehyde for 20 min at room temperature. The lysates were sonicated six times with each time for 30 s using Bioruptor (Diogenode, Denville, NJ, USA). After centrifugation, the supernatant was diluted in ChIP dilution buffer and then incubated overnight at 4 °C with the primary antibody. The eluted DNA was quantified by qPCR and normalized to IgG control.
Confocal microscopy
Immunofluorescence staining and confocal microscopy was used to determine the ATRX (H-300: sc-15408) (Santa Cruz Biotech; 1 : 200), Cdx2 (AB4123) (Chemicon, Billerica, MA, USA; 1 : 200), PML (36-1-104: K0196-3) (MBL, Woburn, MA, USA; 1 : 1000), and trimethyl-Histone H3 (Lys9) (07-442) (Upstate Biotech, Bridgewater, NJ, USA; 1 : 1000) as previously described.5 Images were analyzed using a spinning disk confocal microscope (Olympus DSU, Tokyo, Japan). Deconvolution and 3-dimentainal construction of the confocal image were performed by AQI-X-COMBO-CWF program (Media cybernetics Inc., Bethesda, MD, USA). Control experiments were performed in the absence of primary antibody or in the presence of blocking peptide.
ATRX promoter activity analysis
ATRX-promoter analysis was performed using a reporter-construct containing 5′ regulatory region of the human ATRX gene (−548/+101) including nested deletion constructs (−438/+101; −248/+101). The site-directed mutagenesis was used to generate a mutant reporter with the deletion of Cdx-binding element (Stratagene, Santa Clara, CA, USA). Cells (SH-SY5Y) were transiently transfected with ATRX promoters in the presence or the absence of transcription factors. The luciferase activity was measured 48 h after the transfection. The promoter activity was normalized to protein concentrations.
RNA interference experiment
Cells were transiently transfected with shCdx2 and ATRX RNAi using DMRIE-C transfection reagent (Invitrogen Life Tech., Carlsbad, CA, USA) for 48 h. The sequences of siRNA of ATRX are as following: control siRNA, (forward) 5′-UUCUCCGAACGUGUCACGUTT-3′; (reverse) 5′-ACGUGACACGUUCGGAGAATT-3′; and ATRX siRNA, (forward) 5′-GUGGGCUGAAGAAUUUAAUTT-3′; (reverse) 5′-AUUAAAUUCUUCAGCCCACTT-3′.
Western blot analysis
Cell lysates were prepared from proposed experimental conditions, subjected to SDS-PAGE, and blotted with anti-ATRX (H-300: sc-15408) (Santa Cruz Biotech), anti-Cdx2 (A-22: sc-369 and C-20: sc-583) (Santa Cruz Biotech), and anti- trimethyl-Histone H3 (Lys9) (07-442) (Upstate Biotech) antibody. Protein loading was controlled by probing for β-actin or α-tubulin (Sigma, St. Louis, MO, USA) on the same membrane.
Statistical analysis
The data are presented as the mean±S.E. Data analysis was performed by Student's t-test using StatView 4 (Abacus Concepts, Berkeley, CA, USA). Differences were considered statistically significant when P<0.05.
Abbreviations
- ATRX:
-
alpha thalassemia/mental retardation X linked
- CDX2:
-
caudal type homeobox 2
- ChIP:
-
chromatin immunoprecipitation
- CREB:
-
cAMP response element-binding
- Daxx:
-
death-domain associated protein
- Dnmt3:
-
DNA methyltransferase
- dXNP:
-
drosopila homolog of XNP/ATRX
- GATA2:
-
transcription factors GATA2
- GMR:
-
glass multimer repeat
- HD:
-
Huntington's disease
- H3K9me3:
-
trimethylation of H3K9
- Htt :
-
Huntingtin
- HP1α:
-
heterochromatin protein 1
- MeCP2:
-
methyl-CpG-binding protein 2
- mtHtt:
-
mutant huntingtin
- PML-NBs:
-
promyelocytic leukemia nuclear bodies
- SP1:
-
specificity protein1
- SWI/SNF:
-
switch/sucrose non-fermentable
References
Vonsattel JP, DiFiglia M . Huntington disease. J Neuropathol Exp Neurol 1998; 57: 369–384.
The Huntington’s Disease Collaborative Research Group. A nobel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993; 72: 971–983.
Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA 2000; 97: 6763–6768.
Sugars KL, Rubinsztein DC . Transcriptional abnormalities in Huntington disease. Trends Genet 2003; 19: 233–238.
Ryu H, Lee J, Hargerty SW, Soh BY, Mcalpin SE, Cormier KA et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci USA 2006; 103: 19176–19181.
Sadri-Vakili G, Cha JH . Mechanisms of disease: histone modifications in Huntington's disease. Nat Clin Pract Neurol 2006; 2: 330–338.
Varga-Weisz PD, Becker PB . Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev 2006; 16: 151–156.
Gibbons RJ, Picketts DJ, Villard L, Higgs DR . Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alphathalassemia (ATR-X syndrome). Cell 1995; 80: 837–845.
Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ . ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet 1996; 5: 1899–1907.
Gibbons RJ, McDowell TL, Raman S, O’Rourke DM, Garrick D, Ayyub H et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet 2000; 24: 368–371.
Baumann C, Viveiros MM, De La Fuente R . Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre-implantation embryo. PLoS Genet 2010; 6: e1001137.
Berube NG, Jagla M, Smeenk C, De Repentigny Y, Kothary R, Picketts DJ . Neurodevelopmental defects resulting from ATRX overexpression in transgenic mice. Hum Mol Genet 2002; 11: 253–261.
Ishov AM, Vladimirova OV, Maul GG . Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci 2004; 117: 3807–3820.
Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R et al. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem 2004; 279: 20369–20377.
Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu MJ et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci USA 2007; 104: 2709–2714.
Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ et al. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest 2005; 115: 258–267.
Mangiarini L, Sathasivam K, Seller K, Cozens B, Harper A, Hetherington C et al. Exon1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 1996; 87: 493–506.
Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet 2000; 9: 2799–2809.
McDowell TL, Gibbons RJ, Sutherland H, O’Rourke DM, Bickmore WA, Pombo A et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA 1999; 96: 13983–13988.
Berube NG, Healy J, Medina CF, Wu S, Hodgson T, Jagla M et al. Patient mutations alter ATRX targeting to PML nuclear bodies. Eur J Hum Genet 2008; 16: 192–201.
Taylor JP, Taye AA, Campbell C, Kazemi-Esfarjani P, Fischbeck KH, Min KT . Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev 2003; 17: 1463–1468.
Lee NG, Hong YK, Yu SY, Han SY, Geum D, Cho D . dXNP, a Drosophila homolog of XNP/ATRX, induces apoptosis via Jun-N-terminal kinase activation. FEBS Lett 2007; 581: 2625–2632.
Hong YK, Lee NG, Lee MJ, Park MS, Choi MS, Suh YS et al. dXNP/DATRX increases apoptosis via the JNK and dFOXO pathway in Drosophila neurons. Biochem Biophys Res Commun 2009; 384: 160–166.
Borovecki F, Lovrecic L, Zhou J, Jeong H, Then H, Rosas HD et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci USA 2005; 102: 11023–11028.
Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S et al. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet 2011; 20: 2195–2120.
Mallo GV, Soubeyran P, Lissitzky JC, André F, Farnarier C, Marvaldi J et al. Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J Biol Chem 1998; 273: 14030–14036.
Kazemi-Esfarjani P, Benzer S . Genetic suppression of polyglutamine toxicity in Drosophila. Science 2000; 287: 1837–1840.
Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ . Antioxidants modulate mitochondrial protein kinase A and increase CREB binding to D-Loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci USA 2005; 102: 13915–13920.
Acknowledgements
We thank Dr. Marcy MacDonald for STHdhQ7/7 and STHdhQ111/111 cells and Soung Chan Kim for their technical assistance. This study was supported by NIH NS 067283-01A1 (HR), WCU Neurocytomics Program Grant (800-20080848) (HR), and SRC Grant (2010-0029-403) (HR) from NRF.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by N Bazan
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, J., Hong, Y., Jeon, G. et al. ATRX induction by mutant huntingtin via Cdx2 modulates heterochromatin condensation and pathology in Huntington's disease. Cell Death Differ 19, 1109–1116 (2012). https://doi.org/10.1038/cdd.2011.196
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2011.196
Keywords
This article is cited by
-
Decreased FAK activity and focal adhesion dynamics impair proper neurite formation of medium spiny neurons in Huntington's disease
Acta Neuropathologica (2022)
-
FUS(1-359) transgenic mice as a model of ALS: pathophysiological and molecular aspects of the proteinopathy
neurogenetics (2018)
-
Remodeling of heterochromatin structure slows neuropathological progression and prolongs survival in an animal model of Huntington’s disease
Acta Neuropathologica (2017)
-
Transcription, Epigenetics and Ameliorative Strategies in Huntington’s Disease: a Genome-Wide Perspective
Molecular Neurobiology (2015)