Abstract
The transcription factor p53 is a multifunctional tumor suppressor that arrests the cell cycle in response to stress and modulates the DNA repair process or induces apoptosis. The cellular level and activity of p53 are tightly controlled to maintain proper functioning. This study identified a novel p53-binding glycoprotein, gene related to anergy in lymphocytes (Grail), which formed a negative feedback loop (similar to that of Mdm2). Grail physically and functionally interacted with the N-terminus of p53 to target its degradation and modulate its transactivation activity. Grail also senses and regulates cellular p53 levels, modulates a panel of p53-targeted promoters, and has a role in p53-induced apoptosis in cultured cells. Overexpression of Grail inhibited p53-induced apoptosis by increasing p53 degradation. However, cells not expressing Grail failed to undergo p53-dependent apoptosis, resulting in p21-dependent G1 arrest. Thus, Grail may provide a novel regulatory route for controlling p53 activity under stress conditions.
Similar content being viewed by others
Main
The transcription factor p53 is a multifunctional tumor suppressor involved in cellular responses to various stresses, including DNA damage, hypoxia, low ribonucleotide levels, and oncogene expression.1, 2, 3 Activation of p53 induces cellular responses, such as cell cycle arrest, senescence, and apoptosis, which contribute to its function as a tumor suppressor: either maintaining genomic integrity or eliminating potentially oncogenic cells through apoptosis. The mechanisms that enable p53 to function in these roles involve protein–protein interactions with other key signaling proteins and its ability to bind specific enhancer elements within the promoters of p53 target genes, thereby activating their transcription. However, how p53 selects between genes with different transcriptional programs at the molecular level is not clear. Suppression of p53 activity and that of other tumor suppressors is common in human tumors, in which inactivation of cell-death pathways is one of the central components of progression. The p53 protein is mutated in at least 50% of human tumors. The most frequently observed mechanism underlying p53 inactivation involves point mutations within its DNA-binding domain.4
Gene related to anergy in lymphocytes (Grail; encoded by the RNF128 gene) is a type I transmembrane protein localized in endosomes. This protein is an E3 ubiquitin ligase and is best characterized as a regulator of anergy and cytokine production.5, 6, 7 Activation of the NFATc1 (nuclear factor of activated T-cell 1) homodimer via calcium signaling is responsible for activating the expression of Grail mRNA.8, 9 Grail exists as a tri-molecular complex, comprising Grail, Otub1, and USP8, which controls expression of the Grail protein through the 26S proteasome pathway in anergic T-cells.10, 11, 12, 13 Two recent studies suggest that Grail has roles in other functions besides anergy regulation. The first study investigated the role of Grail in non-lymphoid development, and the other study identified a potential function of Grail in nutrient metabolism.14, 15 The involvement of Grail in regulating the cell cycle and tumorigenesis is unclear.
p21WAF1/Cip1 (later renamed p21) is a well-characterized cyclin-dependent kinase (CDK) inhibitor belonging to the Cip/Kip family.16 This factor mainly inhibits the activity of cyclin/cdk2 complexes and negatively modulates cell cycle progression in the G1 phase.17 As p21 is a transcriptional target for p53, it has a crucial role in mediating growth arrest when cells are exposed to DNA-damaging agents, such as doxorubicin and γ-irradiation.16 Aside from p53, a variety of other factors, including Sp1 (specificity protein 1), p300/CBP, c-Jun, E2F, and Zac1 (zinc-finger protein that regulates apoptosis and cell cycle arrest 1), activate p21 transcription.17, 18, 19, 20, 21 p21 also protects cells against apoptosis independently of cell cycle progression; rather, it regulates gene transcription through multiple protein–protein interactions or through its role in DNA repair. Paradoxically, p21 might also promote apoptosis through both p53-dependent and p53-independent mechanisms under certain cellular stresses.22, 23
The present study identified a p53-interacting glycoprotein, Grail, using the yeast SOS recruitment system,24 and demonstrated that Grail (in addition to Mdm2) is a target for p53, and physically and functionally interacts with the N-terminus of p53 to decrease its protein stability and transactivation activity. In addition, we found that Grail has a role in cell cycle arrest and apoptosis in a p53-dependent manner following treatment with DNA-damaging agents. Thus, this study shows that Grail has a novel, p53-dependent role in regulating the cell cycle and apoptosis.
Results
Grail directly interacts with p53
This study used the newly developed yeast two-hybrid SOS recruitment system to identify novel p53-interacting proteins.24 Of the 1 × 106 individual cdc25–2 yeast transformants screened, 600 colonies were initially isolated from Ynb galactose (leu-, ura-) replica plates incubated at a non-permissive temperature. An additional round of differential growth selection was performed to distinguish galactose- and temperature-dependent transformants from revertants, which yielded nine colonies. Of these colonies, five were false-positive ras clones, one was a p53-independent clone, and three were p53-dependent clones. Sequence analysis revealed that two clones were identical and encoded the carboxyl terminal amino acids of p53 (278–390), and one clone encoded Grail, a 932-bp insert (data not shown).
To validate the interaction between Grail and p53 and to examine whether it was direct or indirect, we performed a GST (glutathione S-transferase) pull-down assay. Purified recombinant GST (negative control) and GST-p53 fusion proteins were used to pull down various in vitro-translated Grail proteins, including Grail1–428 (full length), Grail1–263 (RING domain deleted), and Grail170–428 (PA domain deleted). The results showed that p53 directly interacted with Grail, even in the absence of the RING or PA domains (Figure 1a). In a parallel experiment, we also observed that Grail directly interacted with the N-terminal region of p53 (Figure 1b). Another pull-down assay was performed by adding p53 antibodies to A1-5 (rat embryo fibroblasts transformed with activated ras and a temperature-sensitive mutant of p53) cell extracts with in vitro-synthesized 35S-labeled Grail. The results showed that Grail interacts with p53 in the presence or absence of UV, and the interaction between mdm2 and p53 in the UV irradiation (Figure 1c, compare lanes 1–4). The cell extract from H1299 cells (p53-null) was used as a negative control (Figure 1c, lanes 5 and 6).
Interaction between Grail and p53 in vitro and in vivo. (a and b) Full-length Grail or Grail fragments (a) were translated in vitro and incubated with bead-bound GST or GST-p53. (b) Full-length p53 or p53 fragments were translated in vitro and incubated with bead-bound GST or GST–Grail. Bound proteins were eluted, separated by SDS-PAGE, and subjected to autoradiography. (c) A1–5 and H1299 cell lysates incubated at 37 °C or after treatment with 10 J/m2 UV were subjected to IP with pAb421 and Protein A Sepharose beads. The pulled-down immunocomplexes were incubated with 35S-labeled Grail or Mdm2 as indicated, resolved by SDS-PAGE, and visualized by autoradiography. (d–e) Mouse tissue extracts were prepared from mice treated either with or without IR, followed by IP with a p53 antibody and western blot analysis with a Grail antibody or pre-immune serum. (e) Input of tissues served as the IP control. (f) A549 cell extracts were prepared from cells treated either with or without Act D followed by IP with p53, Grail, or IgG antibody and western blot analysis with Grail or p53 antibody. (NS, non-specific band)
To examine the interaction between Grail and p53 in vivo, cell extracts expressing high levels of Grail (prepared from mouse heart and kidney) were immunoprecipitated with an anti-p53 antibody (pAb421) followed by western blot analysis with antiserum (containing pAb9841) against Grail. The interaction between p53 and Grail was only detected in extracts treated with ionizing radiation (IR) (which induced p53 expression) (Figures 1d and e). Moreover, the interaction was detected in extracts from wild-type mice but not in those from p53-null mice (Supplementary Figure S1). The reciprocal interactions between p53 and Grail were detected in Act D (actinomycin D)-treated A549 cells (Figure 1f). Act D not only induced the protein expression of p53 and Grail, but also enhanced their physical crosstalk.
Grail reduces p53 protein levels
As Grail was identified as an E3 ligase,13 it is possible that Grail degrades p53 via ubiquitination (as does mdm2). To investigate whether Grail affects p53 levels, we used an adenoviral p53 vector to infect human Saos-2 osteogenic cells (p53-null), either with or without the expression of Grail, and examined the levels of p53 protein. The overexpression of Grail significantly reduced the levels of exogenous p53 and its downstream target genes, p21 and Bax (Figure 2a), but this overexpression had no effect on the p53 mRNA levels (Figure 2b). Instead of adenoviral induction, transient expression of Grail remained the ability to reduce the levels of exogenous p53 in Saos-2 cells (Supplementary Figure S2a). Treatment with cycloheximide (CHX), a de novo protein synthesis inhibitor, showed that Grail affected the p53 protein levels by reducing its stability (Figures 2c and d). Furthermore, we found that p53 also reduced the stability of the Grail protein (data not shown). Next, we used the proteasome degradation inhibitor, LLnL, or the lysosome degradation inhibitor, ammonium chloride (NH4Cl), to determine whether Grail degrades p53 in proteasomes or lysosomes. The results showed that neither LLnL nor NH4Cl could rescue the Grail-induced reduction in p53 protein expression (Supplementary Figures S3a and b). LLnL partially stabilized the Grail protein, which was consistent with the characteristics of autoubiquitination.10, 11, 12, 13
Grail reduces p53 protein levels. (a and b) Saos-2 cells were infected with adenoviral p53 and/or Grail. After 48 h, protein (a) and mRNA (b) were extracted and analyzed by western blotting or RT-PCR, respectively. (c and d) Saos-2 cells were infected with adenoviral p53 and/or Grail for 24 h. Next, CHX (100 μg/ml) was added, and cells were harvested at the indicated times. Cell lysates were subjected to western blotting with the indicated antibodies (c). The western blotting data in (d) was quantified using Image J software (NIH, Bethesda, MD, USA). (e) Stable Grail-expressing cell lines, C2C12, and C2C12, were treated with DMSO (control), Act D (30 nM), or UV (200 J/m2) for 48 h. (f) The indicated C2C12 stable cell lines were treated with DMSO (control) or Act D (30 nM) for 48 h followed by western blotting with the indicated antibodies
To determine whether endogenous p53 was downregulated by Grail in response to stress, we established mouse myoblast C2C12 cells that stably overexpresses Grail (C2C12/Grail) and treated them with Act D or UV. After 48 h, p53 protein levels were significantly decreased (Figure 2e, compare lanes 4 and 6 with lane 2). To compare the effects of overexpressing Grail, we established two independent C2C12 cell lines (C2C12/shGrail 1 and 2) that express Grail RNA-mediated short hairpin RNA (shRNA). Under non-stressed conditions, the cells in which Grail was silenced or overexpressed showed low levels of p53 expression. However, p53 only accumulated in C2C12/shGrail cells treated with Act D (Figure 2f, compare lanes 6–9 with lanes 1–4) because Act D did induce Grail mRNA expression in C2C12 cells (Supplementary Figure S2b). The broad Grail bands observed in the Saos-2 cell lysates upon western blot analysis were resolved into narrow 50 kDa bands after treatment with the N-glycosylation inhibitor, tunicamycin (Supplementary Figure S4a), suggesting that Grail is a glycosylated protein. In the presence of Act D and UV, the Grail bands appeared to shift to a higher molecular weight (Figures 2e and f); however, the Act D-induced up-shift was not reversed by tunicamycin treatment (Supplementary Figure S4b), suggesting that Grail contains other, as yet uncharacterized post-translation modifications.
Grail regulates p53 transcriptional activity
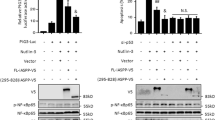
Based on the direct interaction between Grail and p53 (Figures 1a and b), we further investigated whether Grail could regulate the transcriptional activity of p53. First, we examined whether Grail suppressed the transactivation activity of p53 via its N-terminal region (similar to the mechanism of mdm2) (Figure 3). Using a pM vector containing the full-length p53 or the N-terminus of p53 fused to a single Gal4 DNA-binding domain, we observed that the N-terminal region of p53 had a higher transactivation activity than the full-length p53 (Figure 3a, compare histograms 1–3). Both the Grail and Mdm2 proteins suppressed the transactivation activity of the N-terminal region of p53 (Figure 3a, compare histograms 1–9). The combination of Grail and Mdm2 had a greater suppressive effect on both the p53 fusion proteins tested. Second, we examined the repressive effect of Grail on the transcriptional activity of p53 using a 13-copy, p53-responsive element luciferase reporter (pG13-LUC). Overexpression of Grail (and Mdm2) clearly inhibited the reporter activity in the control HeLa/vector cells, whereas both proteins lost the ability to suppress this activity in HeLa cells lacking the expression of p53 (HeLa/shp53 cells) (Figure 3b, left panel). Furthermore, the suppressive effect of both Grail and Mdm2 was restored when exogenous p53 was transfected into the HeLa/vector and HeLa/shp53 cells (Figure 3b, right panel). Next, to examine the effects of Grail on the expression of the pG13-LUC reporter, we constructed two independent C2C12 cell lines in which Grail was stably knocked down by shRNA (C2C12/shGrail 1 and 2). Both cell lines showed increased pG13-LUC reporter activity (Figure 3c, compare histograms 1–4). Next, we examined the effects of Grail on two p53 target genes, p21 and Mdm2, using p53-null non-small lung carcinoma (H1299) cells and quantitative real-time PCR analysis. The expression of exogenous p53 activated both genes in H1299 cells (Figures 3d and e, compare histograms 1 and 2). Consistent with previous observations, the transcriptional activity of p53 on both the p21 and Mdm2 genes was suppressed by Grail in a dose-dependent manner (Figures 3d and e, compare histograms 3–4 with 2).
The effect of Grail on the transactivation activity of p53. (a) HeLa cells were transfected with 0.25 μg of GK1 reporter DNA and 0.25 μg of pM.p53 fl (full length) or pM.p53N (N-terminal fragment) in the presence of 0.25 μg of Grail, Mdm2, or both. The luciferase activity in the transfected cell extracts was subsequently determined. (b) HeLa/vector cells (open columns) and HeLa/p53 shRNA cells (closed columns) were transfected with 0.25 μg of pG13-LUC reporter DNA and 0.25 μg of Grail or Mdm2. The luciferase activity in the transfected cell extracts was later determined. (c) C2C12/vector, C2C12/shp53, C2C12/shGrail 1, and C2C12/shGrail 2 cells were transfected with 0.25 μg of pG13-LUC reporter DNA. The luciferase activity in the transfected cell extracts was subsequently determined. The luciferase activity in the transfected cell extracts was then determined. (d and e) The endogenous p21 and MDM2 gene expression level was later determined. (d and e) H1299 cells were transfected with indicated amounts of Grail in the presence of 100 ng of p53 for 24 h, and the mRNA amount was analyzed by quantitative real-time PCR. Data (a–d) represent the mean±S.D. from three separate experiments
Grail is a putative p53 target gene
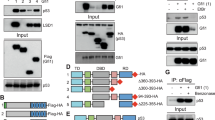
Based on the working model of the interaction between Mdm2 and p53,25 we further examined whether Grail is a p53 target gene. We identified two putative p53-responsive elements in the promoter region of the Grail gene (Supplementary Figure S5). DAPAs (DNA-affinity precipitation assays) were performed using the p53-responsive element of p21 as a positive control (Figure 4a). Using two mutant p53-responsive elements from Grail, we identified site 2 as the primary p53-responsive element (Figure 4b). Renal cell carcinoma 789-0/shp53 cells lacking the expression of endogenous p53 showed a significant reduction in Grail expression (Figure 4c). This result was further confirmed by ChIP (chromatin immnuoprecipitation) analysis, which demonstrated that p53 did indeed interact directly with sites 1 and 2 of the Grail promoter (Figure 4d). Many p53 target genes respond differently to different amounts of p53; therefore, we examined the dose effect of p53 on the Grail promoter compared with that of other promoters (PUMA, Mdm2, p21, and Bax) (Figure 4e). The results showed that the p53 dose-response pattern to the Grail promoter was similar to that of PUMA and p21. Furthermore, we used a maximal dose of p53 to examine the effect of Zac1, a coactivator of p53, on the Grail promoter.26 Our data showed that Zac1 acts synergistically with p53 on the promoters for all of the tested p53 target genes (Figure 4f).
Grail is a p53 target gene. (a and b) H1299 cells were infected with adenoviral p53 for 24 h and the cell extracts collected. (a) Two potential Grail p53-responsive elements (1 and 2) (lanes 3 and 4), a positive control (p53-responsive element of p21, lane 2), and a negative control (lane 5). (b) Positive control (p53-responsive element of p21, lane 1), wild-type, and mutants of the potential p53-responsive elements 1 and 2 of Grail (lanes 2–5). Biotin-labeled probes were used to perform the DAPA analysis. (c) RCC 786-O cells were silenced for endogenous p53 expression, and the silencing efficiency was checked by RT-PCR and western blot analysis. (d) RCC 786-O/vector and/shp53 cells were used for ChIP analysis as described in the methods section. The eluted DNAs were analyzed by PCR. (e and f) HeLa cells were transiently transfected with 0.25 μg of the indicated p53 target promoters, (e) the indicated amounts of p53, or (f) a specific amount of p53, as well as 0.25 μg pSG5.HA.Zac1. The luciferase activity in the transfected cell extracts was later determined
Grail suppresses stress-induced apoptosis
The tumor suppressor p53 has an important role in regulating cell cycle arrest and apoptosis.1 Therefore, we further examined the effects of Grail on the function of p53 during cell cycle arrest and apoptosis. The DNA profiles of a pair of C2C12 cell lines expressing endogenous wild-type p53 (C2C12 and C2C12/Grail) in response to various stresses, such as Act D and UV, were examined by FACS (fluorescence-activated cell sorting) analysis (Figure 5). We demonstrated that the percentage of C2C12 cells undergoing apoptosis after treatment with Act D or UV was significantly lower when the cells overexpressed Grail (C2C12/Grail) than it was in the parental line (reduced from 56 to 29% versus 45 to 23% for Act D versus UV, respectively) (Figure 5a). To confirm the effect of Grail on p53-induced apoptosis, we next analyzed the anti-apoptotic effects of Grail in Saos-2 cells. Consistent with the effects shown in Figure 5a, the overexpression of Grail suppressed p53-induced apoptosis by ∼32% at 48 h after p53 induction (Figure 5b). One of the most important questions to answer regarding the anti-apoptotic function of Grail overexpression is whether it can inhibit apoptosis in the absence of p53. To this end, we established stable C2C12/shp53 (p53-silenced) and C2C12/shp53/Grail (p53-silenced, Grail-overexpressing) cell lines. The result clearly showed that apoptosis was unaffected by the expression of Grail in these cell lines (Figure 5c). We also overexpressed Grail in Saos-2 cells and subsequently treated them with UV. These data indicated that Grail does not affect apoptosis in the absence of p53 (Figure 5d). Taken together, these data suggest that overexpression of Grail decreases the stress-induced p53 abundance and, subsequently, decreases the p53-induced apoptotic effect.
The anti-apoptotic effects of Grail. (a) C2C12- and C2C12-Grail stable cell lines were treated with DMSO (control), Act D (30 nM), or UV (200 J/m2) for 48 h. The cells were later harvested and cell cycle profiles were analyzed by FACS. (b) Saos-2 cells were infected with adenoviral p53 and/or Grail for 48 h. The cells were harvested and the cell cycle profiles were analyzed by FACS. (c) C2C12/shp53- and C2C12/shp53/Grail-stable cell lines were treated with DMSO (control), Act D (30 nM), or UV (200 J/m2) for 48 h. The cells were harvested, and the cell cycle profiles were analyzed by FACS. (d) Saos-2 cells (with or without Grail expression) were treated with UV (200 J/m2) for 48 h as indicated. Cells were subsequently harvested, and the cell cycle profiles were analyzed by FACS
To further investigate the anti-apoptotic effects of Grail, the DNA profiles of the C2C12 and C2C12/shGrail 2 cell lines were examined in the absence and presence of various stresses by FACS. Surprisingly, the percentage of apoptotic C2C12/shGrail 2 cells was significantly reduced after Act D or UV treatment (from 56 to 8% and from 45 to 16%, respectively) (Figure 6a and b). We observed a similar response after overexpressing or silencing Grail in U2OS cells, which is another cell line that expresses functional p53 (Supplementary Figures S6a and b). To analyze further the effects of silencing Grail on p53-dependent apoptosis induced after DNA damage, we established Grail-silenced Saos-2 (Saos-2/shGrail) cell lines and analyzed the cell cycle profiles after adenoviral p53 infection and UV treatment. Apoptosis was observed in the Saos-2 cells infected with adenoviral p53 or treated with UV (Supplementary Figures S7a and b). However, apoptosis was reduced to a greater extent in Saos-2/shGrail cells infected with p53 (from 55 to 14%) compared with that in Saos-2/shGrail cells treated with UV (from 42 to 30%), implying that Grail has an important role in p53-induced apoptosis.
Downregulation of Grail inhibits p53-mediated apoptosis and depends on p21. (a) C2C12 cells stably expressing the indicated shRNAs were treated with DMSO (control), Act D (30 nM), or UV (200 J/m2). After 48 h, cells were harvested and the cell cycle profiles were analyzed by FACS. (b) Efficiency of Grail silencing in C2C12 cells was analyzed by RT-PCR. (c) Efficiency of silencing p21 in C2C12 cells as analyzed by RT-PCR. (d) C2C12 stably silenced p21 cells were treated with DMSO (control) or Act D (30 nM). After 48 h, the cells were harvested, and the cell cycle profiles were analyzed by FACS
Grail acts as a switchbox for stress-induced apoptosis and cell cycle arrest via a p21-dependent pathway
This study has shown that in the absence of Grail, p21 is upregulated after DNA damage (Figure 2f, compare histograms 6 and 1 or 7 and 2). We also found that the G1 peak became more obvious in parallel with the reduction in the number of cells in sub-G1 (Figure 6a and Supplementary Figure S7a) under stressed conditions. However, in the absence of Grail, a robust G1 arrest may occur due to the upregulation of p21 after DNA damage. Therefore, we next investigated the role of p21 and its functional correlation with the loss of Grail during stress-induced apoptosis. We established p21-silenced C2C12 cell lines and examined the silencing efficacy by western blot analysis (Figure 6c). We also established two stable double-silenced C2C12/shGrail/shp21 cell lines. As shown in Figure 6d, the downregulation of Grail expression (C2C12/shGrail cells) inhibited DNA damage-induced apoptosis (most cells are in G1 phase); however, no switch from sub-G1 to G1 was observed in the C2C12/shGrail/shp21 cells. These findings suggested that Grail prevents cells from arresting at the G1 phase via a p21-dependent pathway in response to DNA damage.
To examine whether the expression of p53 target genes, PUMA and Grail, is correlated with the observation of apoptosis induced by p53 in cells null for p53, we infected H1299 cells with adenoviruses overexpressing p53. Our flow-cytometry and quantitative real-time PCR data demonstrated that p53 induced the levels of the sub-G1 stage (12 h: 5% and 24 h: 21%), as well as the expression of the apoptosis-related PUMA gene (relative control mRNA expression fold at 12 h: 4.4; and at 24 h: 14) (Figures 7a and b). Our previous data revealed that Grail was a p53 target gene and had an important role in the p53-dependent apoptosis pathway (Figure 4 and Supplementary FigureS7). In this experiment, our data confirmed that Grail was induced by p53 (relative control mRNA expression fold at 12 h: 15; and at 24 h: 32) and coincidentally expressed with PUMA in the p53-dependent apoptosis (Figures 7a and b).
The p53/Grail negative feedback loop. (a and b) H1299 cells were infected with Ad-p53 expression vector. At indicated time, cells were harvested to analyze the cell cycle profiles by FACS (a), and analyze the mRNA amount by quantitative real-time PCR (b). (c and d) U2OS cells stably expressing the indicated shRNAs were treated with DMSO (control) or Act D (20 nM) for 24 h. Representative immunofluorescence images showing triple labeling for Grail (green), p53 (red), DAPI (blue), and the merged images of Grail/p53 and Grail/p53/DAPI
Finally, we examined the subcellular localization of endogenous p53 and Grail proteins after Act D treatment or silencing the p53 or Grail gene expression using immunofluorescence microscopy in U2OS cells. In Figure 7c, Act D induced the accumulation of nuclear p53 proteins and also increased the abundance of cytoplasmic and nuclear Grail protein. Silencing the endogenous p53 gene not only reduced the p53 protein expression but also Grail protein expression, regardless of whether U2OS cells were treated with Act D. We further silenced endogenous Grail gene expression and found that endogenous p53 proteins were induced (Figure 7d).Therefore, our findings were consistent with our previous understanding: Grail is a p53 target gene, and it is involved in the stability of p53.
Discussion
This study identified a p53-interacting glycoprotein, Grail, which, similar to Mdm2, is also a p53 target gene and is involved in the stability of p53. Grail not only physically interacts with p53 but also functionally links to p53 to modulate its cellular effects. Our data suggest that Grail is required by cells undergoing p53-induced apoptosis in response to various stresses (Figures 5 and 6 and Supplementary Figure S7); however, this gene’s overexpression suppresses p53-induced apoptosis by targeting p53 for degradation. Overexpression of Grail does not affect the cell cycle progression in unstressed cells. The downregulation of Grail expression causes p53-induced apoptosis to switch to p21-dependent G1 arrest under various stressed conditions, and suggests that Grail has an essential role in determining the cell fate (apoptosis or cell cycle arrest). Such cell fate determination depends on the binding affinity of the p53 transcription factor to its target genes, such as p21, which regulates cell cycle arrest and Puma, which regulates pro-apoptotic responses. Grail, as well as p21 and PUMA, is a p53 target gene, and its p53 responsive site is more sensitive than other targets in our analytical conditions (Figures 4e and f). Therefore, we propose a working model based on our findings (Figure 8): cellular p53 transcription factor is activated and translocated into the nucleus by DNA damage stresses and transcribes its target genes, including Grail, PUMA, and p21. The p53/Grail negative feedback loop not only regulates the abundance of p53 and Grail (pathway I) but also initiates p53-dependent apoptosis (pathway II). One possibility is that the cellular level of Grail protein is the primary factor determining the functional role of p53 in cell cycle arrest (via p21) or apoptosis (via PUMA) under stressed conditions. Our unpublished data showed that expression of Grail might be downregulated through epigenetic modifications in oral cancers, suggesting that this could provide a growth advantage during tumorigenesis. Cancer cells expressing relatively low levels of Grail can overcome DNA stress by arresting at G1 stage, rather than undergoing cellular apoptosis. In contrast to the p53/Mdm2 negative feedback loop, the blockade of MDM2 results in p53 accumulation and the induction of p53-dependent cell cycle arrest and apoptosis. The detailed crosstalk between Grail and Mdm2 for the regulation of p53 functions remains to be investigated.
The working model of Grail in the regulation of p53-dependent functions. In normal conditions, the endogenous p53 abundance is too low to transcribe its target genes. DNA damage induces endogenous p53 expression sufficiently to transcribe its target genes, which include p21, PUMA, and Grail. In pathway I, increasing the Grail proteins are able to degrade stress-activated p53 proteins. In pathway II, the p53/Grail negative feedback loop is required for apoptosis induction. The downregulation of Grail gene expression and protein, via epigenetic modifications, fails to maintain the p53-dependent apoptosis. The stressed cells later enter the p21-dependent cell cycle G1 arrest, due to increased endogenous p53 proteins. Further silencing of the p21 gene results in the predominance of endogenous Grail proteins for the p53-dependent apoptosis. Therefore, Grail is a key protein for the regulation of p53 mediated functions in apoptosis and cell cycle G1 arrest
Studies show that the subcellular localization of p21 influences its activity during the apoptotic process.27, 28 p21 in the cytoplasm blocks the activation of pro-caspase 3, thereby inhibiting apoptosis, whereas the p21 in the nucleus promotes its degradation and creates conditions that favor apoptosis in response to DNA damage. To date, it is still unclear which regulators are responsible for controlling p53-mediated cell cycle arrest and apoptosis. Our studies show that Grail interacts with p53 to degrade endogenous or damage-induced p53 proteins under both unstressed and stressed conditions. Grail may regulate cellular p21 functions via indirect (the competition of p53-dependent transactivation) or direct (the suppression of p21-dependent G1 arrest) pathways. In the stressed condition, Grail is transcribed by DNA-damage-induced p53 via a negatively feedback mechanism. This mechanism regulates the level of endogenous p53 proteins involved in the p53-dependent apoptosis and p21-dependent G1 arrest. As Grail is shown to be downregulated in a subset of human tumors (data not shown), it is proposed that Grail represents a means by which p53 activity can be attenuated in cancer. Through the epigenetic modifications, reduced Grail expression loses the ability to compete with p21 and change cell fate into G1 arrest. However, further downregulation of p53-induced p21 levels enables endogenous Grail to induce apoptosis. Therefore, our study indicates that Grail is a key player to determine the cell fate in the p53-dependent apoptosis and cell cycle arrest.
In addition to TCR activation in human T-cells, the hematopoiesis pathway, and the maintenance of liver glucose and lipid metabolism,5, 9, 14, 15 Grail also has a functional role in determining p53-dependent cell fate. In Rnf128−/− cells, TCR activation leads to substantially increased expression of NFATc1 mRNA and protein.9 Similar to the hypermethylation of the Grail promoter, NFATc1 was also reported to undergo this type of epigenetic modification in human lymphomas with defects in immunoreceptor signaling.29 It is possible that Grail might be involved in crosstalk between NFAT and p53 during cancer development. A recent finding shows that endogenous Grail regulates cell cycle entry and promotes the proliferation of primary naïve CD4 T-cells;10 therefore, it would be interesting to examine the possibility that p53 is required for Grail functions during T-cell unresponsiveness.
Experimental Procedures
Cell culture, plasmids, and transfection
A1-5, Saos-2, H1299, A549, C2C12, HeLa, and RCC 789-0 cells were cultured in Dulbecco modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (Invitrogen, Carlsbad, CA, USA). Wild-type Grail and wild-type p53 were cloned into pCDNA3 vectors (Invitrogen) via the BamHI–NotI and EcoRI–NotI sites, respectively. Full-length and various truncated Grail DNAs were cloned into the EcoRI–NotI sites of the pCMV.TNT vector (Promega, Madison, WI, USA). Bacterial expression vectors for various GST-p53 or -Grail fusions were constructed by inserting the appropriate PCR fragments into the EcoRI–XhoI sites of pGEX-4T1 (GE HealthCare, Waukesha, WI, USA). Reporters for pG13-, p21-, Mdm2-, PUMA-, and Bax-LUC have been described previously.26, 30
Cell transfections were performed using Fugene 6 (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Cells were plated at a low density (∼1 × 105 cells/60 mm dish) and allowed to grow to 30–40% confluence, at which time various genes were transfected as indicated using Fugene 6. The transfected cells were harvested at 48 h and lysed in RIPA buffer (100 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1% SDS, and 1% Triton X-100).
Protein–protein interaction assays
Coupled in vitro transcription and translation reactions were performed using the TNT T7 system following the manufacturer’s instructions (Promega). For the GST pull-down assays, 35S-labeled proteins were produced using the TNT T7-coupled reticulocyte lysate system (Promega) and GST fusion proteins were produced in Escherichia coli, BL21. Various radioactively labeled fragments of Grail or p53 protein were translated in vitro, followed by incubation with the indicated immobilized GST–p53 or GST–Grail fusion proteins. The products were eluted and analyzed by SDS-PAGE. For the in vitro binding assays, cell extracts (prepared from A1-5, UV-treated A1-5 or H1299 cells) were mixed with pAb421 and Protein A Sepharose in lysis buffer containing protease inhibitors at 4 °C for 1 h. After a brief wash with SNNTE buffer, 30 μl of translated proteins (35S-labeled mouse Grail, p53 or mdm2) were added to the immune complexes and incubated for 30 min at 30 °C. At the end of the incubation, the binding complexes were subjected to SDS-PAGE after three brief washes with SNNTE buffer. The gel was fixed with 10% acetic acid and 30% methanol, treated with a solution containing 1 M sodium salicylate and 5% glycerol, dried and exposed to an X-ray film for 24 h.
Immunoprecipitation and western blot analysis
Cells were harvested in lysis buffer (50 mM Tris, pH 8.0, 5 mM NaCl, 0.5% NP-40 and 1 × protease inhibitor), freeze/thawed three times, and the protein recovered. The protein concentration was determined using the Bradford method (Bio-Rad, Hercules, CA, USA). Cell extracts containing equivalent amounts of protein were immunoprecipitated in lysis buffer containing a monoclonal antibody against p53 overnight at 4 °C. Protein A Sepharose beads were added to the immunoprecipitation mixture for 1 h before three washes with SNNTE buffer (5% sucrose, 1% NP-40, 0.5 M NaCl, 50 mM Tris, pH 7.4, and 5 mM EDTA). The entire immunoprecipitate was then suspended in SDS-PAGE sample buffer, boiled, and loaded onto an SDS-polyacrylamide gel. The separated proteins were transferred to a nitrocellulose membrane and the blot was probed with the indicated primary antibodies followed by a secondary antibody (horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG in PBS/Tween 20 with 5% Carnation non-fat milk). Proteins were detected using enhanced chemiluminescence reagents (GE HealthCare). The primary antibodies used for immunoblotting were: monoclonal anti-p53 (pAb421), anti-actin (MAb1501; Chemicon, Temecula, CA, USA), anti-actinin (H-2; Santa Cruz Biotechnology, Santa Cruz, CA, USA); polyclonal anti-Grail, and anti-p21 (C-19; Santa Cruz Biotechnology).
Luciferase reporter assays
Luciferase assays were performed using the Promega Luciferase Assay Kit. The total DNA used for reporter analysis was adjusted to 1 μg by adding the necessary amount of empty vector. The luciferase activity of the transfected cell extracts was presented as relative light units (RLU) and expressed as the mean±S.D. of three transfected cultures.
DAPA
PCR products were labeled with biotin (biotin 3′-labeling Kit, PIERCE, Rockford, IL, USA). Oligonucleotide primers specific for p53 responsive element 1 (forward: 5′- GAACTGAAAAGTGGCTAAGAC -3′ and reverse: 5′- TGTAATAAAGCACTTAGAACAATG -3′; deletion region: −1278/−1305) and p53 responsive element 2 (forward: 5′- GGCAGAAGGGAATCGTGA -3′ and reverse: 5′- TTTCTTGTATAAACCACCAACTAG -3′; deletion region: +9304/+9332) were used in these experiments. H1299 nuclear extracts were incubated with DAPA binding buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, and 1 mM DTT) as described previously.30 Specific protein–DNA–agarose complexes were washed, analyzed by SDS-PAGE, and detected by immunoblotting with an anti-p53 antibody.
Chromatin IP (ChIP) assay
ChIP assays were performed in RCC 789-0/vector or RCC 789-0/shp53 cells. Cells were crosslinked with 1% formaldehyde at 37 °C for 15 min as described previously.31 IP was performed with 1 μg of anti-p53 or anti-normal mouse IgG with rotation at 4 °C overnight. The following day, the chromatin–antibody complexes were eluted from the solution by incubating with 40 μl of salmon sperm DNA-saturated 50% protein A/G-Sepharose at 4 °C for 2 h. The beads were harvested and washed as described above. Crosslinking was reversed by heating at 65 °C overnight followed by treatment with 100 μg/ml proteinase K at 50 °C for 1 h. DNA was extracted with a gel extraction kit (Geneaid, Taiwan, ROC) and dissolved in 60 μl water. Primer pairs specific for p53-responsive elements 1 and 2 of the Grail promoter region were used to amplify the positive control (responsive element 1 forward: 5′- GAACTGAAAAGTGGCTAAGAC -3′ and reverse: 5′- TGTAATAAAGCACTTAGAACAATG -3′; responsive element 2 forward: 5′- GGCAGAAGGGAATCGTGA -3′ and reverse: 5′- TTTCTTGTATAAACCACCAACTAG -3′).
FACS analysis
Analysis was based on the measurement of the DNA content of cell nuclei labeled with propidium iodide. To evaluate the cell cycle profiles, cells were treated as for the proliferation experiments, washed with ice-cold PBS, and incubated with propidium iodide (0.05% in PBS, 0.1% Triton X-100, and 0.01% RNase) for 15 min at room temperature in the dark. Cells were subsequently analyzed using a FACS Calibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA), and data were analyzed using Cell Quest Pro software (BD Biosciences).
Virus production, viral transduction, and RNA interference
Two adenoviruses, adeno-p53 and adeno-Grail, were constructed using the Adeno-X Expression System developed by BD Clontech (Mountain View, CA, USA). Adenoviruses were prepared in accordance with standard protocols (Clontech). Grail was cloned into the retroviral plasmid vector, pQCXIP (Clontech). The pQCXIP–Grail and pQCXIP–empty plasmids were later transfected into GP2-293 cells using Fugene 6 (Roche). Grail retrovirus was prepared according to the protocol published on the Clontech website (Grail shRNA 2 target sequence: 5′- GCAGGAAGCAGAGGCAGTTAA -3′; Grail shRNA 4 target sequence: 5′- GGAGCAGATGAGCCACCTCTG -3′; p53 shRNA target sequence: 5′- GGAGGAGTCACAGTCGGATAT -3′). The shRNA oligonucleotides were cloned into the retroviral siRNA expression vector, pSIREN-Retro-Q (Clontech). Retroviruses overexpressing Grail shRNA and p53 shRNA were generated according to the protocol published on the Clontech website. P21-shRNA-containing lentiviral vectors were purchased from the National RNAi Core Facility (Academia Sinica, Taiwan, ROC) and prepared in accordance with standard protocols. Cells were infected with the indicated retroviruses or lentiviruses in selection medium containing 2 μg/ml polybrene. Forty-eight hours after infection, cells were treated with 8 μg/ml puromycin to select for a pool of puromycin-resistant clones.
RT-PCR and quantitative RT-PCR
Total RNA was isolated using the TRIZol reagent (Sigma, St. Louis, MO, USA). The OneStep RT-PCR Kit (Qiagen) was used for RT-PCR and reactions were run on a GeneAmp PCR system 9700 (Applied Biosystems, Hammonton, NJ, USA). The following primers were used for RT-PCR: Grail forward: 5′- CATGGGGCAAGTCCTGTGTCCTAA -3′ and reverse: 5′- ACGTGTCCTGGCGGGTTCC -3′; p53 forward: 5′- CAGTCTGGGACAGCCAAGTC -3′ and reverse: 5′- CTTCTGTACGGCGGTCTCTC -3′; GAPDH forward: 5′- AGCCAAAAGGGTCATCATCTC -3′ and reverse: 5′- GTCCACCACCCTGTTGCTGTAG -3′. Gene expression was examined with the Applied Biosystems 7500 Real-Time PCR System using IQ2 FAST qPCR kit. The following primers were used for quantitative real-time PCR: Grail forward: 5′- GCGTCTGGAGCCGTCATCTTTA -3′ and reverse: 5′- GGGCCATGTTTTTTCCCTACTTCTAT -3′; p21 forward: 5′-GAGAGCGGCGGCAGACAACAGG-3′ and reverse: 5′- GCGCCCAATACGACCAAATC -3′; mdm2 forward: 5′- GGCTCTGTGTGTAATAAGGGAGA -3′ and reverse: 5′- GGACTGCCAGGACTAGACTTTG -3′; PUMA forward: 5′- ATGGCGGACGACCTCAAC -3′ and reverse: 5′- GTATGCTACATGGTGCAGAGAAAG -3′; GAPDH forward: 5′- AGCCAAAAGGGTCATCATCTC -3′ and reverse: 5′- GTCCACCACCCTGTTGCTGTAG -3′.
Immunofluorescence staining
The cells were grown on glass coverslips under DMSO or Act D (20 nM) during 24 h. Cells were later fixed in 4% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X-100 for 10 min. The slides were washed three times with PBS and blocked with 2% BSA in PBS for 30 min, and incubated with the primary antibody rabbit polyclonal anti-Grail or mouse monoclonal anti-p53 (DO-1) overnight at 4 °C in 1% BSA in PBS, and later with the secondary Alexa-Fluor 488 or Alexa-Fluor 594 antibody (Invitrogen) for 1 h in 1% BSA in PBS at 37 °C, and the nuclei were stained with DAPI for 10 min. Cells were mounted with vectashield (Vector Laboratories, Barlingame, CA, USA), and images were acquired on a Leica DMI 6000B fluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Statistical analysis
One-way analysis of variance (ANOVA) was used to assess the differences in the means between groups, and Student’s two-tailed t-test was used to determine differences between the means of any two groups. A P value of <0.05 was regarded as statistically significant.
Abbreviations
- Grail:
-
gene related to anergy in lymphocytes
- NFATc1:
-
nuclear factor of activated T-cell 1
- Sp1:
-
specificity protein 1
- Zac1:
-
zinc-finger protein that regulates apoptosis and cell cycle arrest 1
- Act D:
-
actinomycin D
- UV:
-
ultraviolet
- IR:
-
ionizing radiation
- GST:
-
glutathione S-transferase
- CHX:
-
cycloheximide
- shRNA:
-
short hairpin RNA
- DAPA:
-
DNA-affinity precipitation
- ChIP:
-
chromatin immnuoprecipitation
- FACS:
-
fluorescence-activated cell sorting
- RT-PCR:
-
reverse transcriptase-polymerase chain reaction
- SDS-PAGE:
-
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
References
Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y . Tumour suppression by p53: the importance of apoptosis and cellular senescence. J Pathol 2009; 219: 3–15.
Jiang L, Sheikh MS, Huang Y . Decision making by p53: life versus death. Mol Cell Pharmacol 2010; 2: 69–77.
May P, May E . Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 1999; 18: 7621–7636.
Menendez D, Inga A, Resnick MA . The expanding universe of p53 targets. Nat Rev Cancer 2009; 9: 724–737.
Anandasabapathy N, Ford GS, Bloom D, Holness C, Paragas V, Seroogy C et al. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity 2003; 18: 535–547.
Seroogy CM, Soares L, Ranheim EA, Su L, Holness C, Bloom D et al. The gene related to anergy in lymphocytes, an E3 ubiquitin ligase, is necessary for anergy induction in CD4 T cells. J Immunol 2004; 173: 79–85.
Whiting CC, Su LL, Lin JT, Fathman CG . GRAIL: a unique mediator of CD4 T-lymphocyte unresponsiveness. FEBS J 2011; 278: 47–58.
Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K et al. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol 2004; 5: 255–265.
Nurieva RI, Zheng S, Jin W, Chung Y, Zhang Y, Martinez GJ et al. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity 2010; 32: 670–680.
Lin JT, Lineberry NB, Kattah MG, Su LL, Utz PJ, Fathman CG et al. Naive CD4 T cell proliferation is controlled by mammalian target of rapamycin regulation of GRAIL expression. J Immunol 2009; 182: 5919–5928.
Su LL, Iwai H, Lin JT, Fathman CG . The transmembrane E3 ligase GRAIL ubiquitinates and degrades CD83 on CD4 T cells. J Immunol 2009; 183: 438–444.
Lineberry N, Su L, Soares L, Fathman CG . The single subunit transmembrane E3 ligase gene related to anergy in lymphocytes (GRAIL) captures and then ubiquitinates transmembrane proteins across the cell membrane. J Biol Chem 2008; 283: 28497–28505.
Soares L, Seroogy C, Skrenta H, Anandasabapathy N, Lovelace P, Chung CD et al. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat Immunol 2004; 5: 45–54.
Nakamichi S, Senga Y, Inoue H, Emi A, Matsuki Y, Watanabe E et al. Role of the E3 ubiquitin ligase gene related to anergy in lymphocytes in glucose and lipid metabolism in the liver. J Mol Endocrinol 2009; 42: 161–169.
MacKenzie DA, Seroogy CM . Sustained expression of GRAIL during hematopoiesis results in dysregulated differentiation. Acta Haematol 2009; 122: 230–237.
el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825.
Gartel AL, Najmabadi F, Goufman E, Tyner AL . A role for E2F1 in Ras activation of p21(WAF1/CIP1) transcription. Oncogene 2000; 19: 961–964.
Billon N, Carlisi D, Datto MB, van Grunsven LA, Watt A, Wang XF et al. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 1999; 18: 2872–2882.
Kardassis D, Papakosta P, Pardali K, Moustakas A . c-Jun transactivates the promoter of the human p21(WAF1/Cip1) gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J Biol Chem 1999; 274: 29572–29581.
Koutsodontis G, Tentes I, Papakosta P, Moustakas A, Kardassis D . Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J Biol Chem 2001; 276: 29116–29125.
Liu PY, Chan JY, Lin HC, Wang SL, Liu ST, Ho CL et al. Modulation of the cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by Zac1 through the antagonistic regulators p53 and histone deacetylase 1 in HeLa Cells. Mol Cancer Res 2008; 6: 1204–1214.
Pang LY, Scott M, Hayward RL, H Mohammed, Whitelaw CB, Smith GC et al. p21(WAF1) is component of a positive feedback loop that maintains the p53 transcriptional program. Cell Cycle 2011; 10: 932–950.
Abbas T, Dutta A . p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9: 400–414.
Huang W, Wang SL, Lozano G, de Crombrugghe B . cDNA library screening using the SOS recruitment system. Biotechniques 2001; 30: 94–98 100.
Momand J, Wu HH, Dasgupta G . MDM2-master regulator of the p53 tumor suppressor protein. Gene 2000; 242: 15–29.
Huang SM, Schonthal AH, Stallcup MR . Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1. Oncogene 2001; 20: 2134–2143.
Asada M, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K et al. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J 1999; 18: 1223–1234.
Didenko VV, Wang X, Yang L, Hornsby PJ . Expression of p21(WAF1/CIP1/SDI1) and p53 in apoptotic cells in the adrenal cortex and induction by ischemia/reperfusion injury. J Clin Invest 1996; 97: 1723–1731.
Akimzhanov A, Krenacs L, Schlegel T, Klein-Hessling S, Bagdi E, Stelkovics E et al. Epigenetic changes and suppression of the nuclear factor of activated T cell 1 (NFATc1) promoter in human lymphomas with defects in immunoreceptor signaling. Am J Pathol 2008; 172: 215–224.
Huang SM, Huang SP, Wang SL, Liu PY . Importin alpha1 is involved in the nuclear localization of Zac1 and the induction of p21(WAF1/CIP1) by Zac1. Biochem J 2007; 402: 359–366.
Ma H, Shang Y, Lee DY, Stallcup MR . Study of nuclear receptor-induced transcription complex assembly and histone modification by chromatin immunoprecipitation assays. Methods Enzymol 2003; 364: 284–296.
Acknowledgements
This work was supported by grants from the National Science Council (NSC 98–2320–B–016–012–MY3 to S-M Huang) and Teh-Tzer Study Group for Human Medical Research Foundation (A100–1032 to C-L HO), Taiwan, ROC.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Oren
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, YC., Chan, JH., Chiu, YL. et al. Grail as a molecular determinant for the functions of the tumor suppressor p53 in tumorigenesis. Cell Death Differ 20, 732–743 (2013). https://doi.org/10.1038/cdd.2013.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2013.1
Keywords
This article is cited by
-
RNF128 regulates neutrophil infiltration and myeloperoxidase functions to prevent acute lung injury
Cell Death & Disease (2023)
-
p53 regulation by ubiquitin and ubiquitin-like modifications
Genome Instability & Disease (2022)
-
Requirement of splicing factor hnRNP A2B1 for tumorigenesis of melanoma stem cells
Stem Cell Research & Therapy (2021)
-
E3 ubiquitin ligase Grail promotes hepatic steatosis through Sirt1 inhibition
Cell Death & Disease (2021)
-
Downregulation of RNF128 activates Wnt/β-catenin signaling to induce cellular EMT and stemness via CD44 and CTTN ubiquitination in melanoma
Journal of Hematology & Oncology (2019)