Abstract
Neuroprotectin D1 (NPD1), a docosahexaenoic acid (DHA)-derived mediator, induces cell survival in uncompensated oxidative stress (OS), neurodegenerations or ischemic stroke. The molecular principles underlying this protection remain unresolved. We report here that, in retinal pigment epithelial cells, NPD1 induces nuclear translocation and cREL synthesis that, in turn, mediates BIRC3 transcription. NPD1 activates NF-κB by an alternate route to canonical signaling, so the opposing effects of TNFR1 and NPD1 on BIRC3 expression are not due to interaction/s between NF-κB pathways. RelB expression follows a similar pattern as BIRC3, indicating that NPD1 also is required to activate cREL-mediated RelB expression. These results suggest that cREL, which follows a periodic pattern augmented by the lipid mediator, regulates a cluster of NPD1-dependent genes after cREL nuclear translocation. BIRC3 silencing prevents NPD1 induction of survival against OS. Moreover, brain NPD1 biosynthesis and selective neuronal BIRC3 abundance are increased by DHA after experimental ischemic stroke followed by remarkable neurological recovery. Thus, NPD1 bioactivity governs key counter-regulatory gene transcription decisive for retinal and brain neural cell integrity when confronted with potential disruptions of homeostasis.
Similar content being viewed by others
Main
Cell survival is tightly regulated in the nervous system to facilitate selective cell removal during development to ensure proper neuronal circuit integrity and throughout life as a response to potential disruptions of homeostasis. Thus, neural cells have the ability to activate pathways that crosstalk one to another, and then assemble consolidated responses that decide cell fate.
Nuclear factor kappa B (NF-κB) activation embraces a summation of stimuli that converge in cell fate decisions with the formation of three types of complexes: Complex I, which activates NF-κB; Complex II, which leads to apoptosis; and Complex III, which results in necrosis.1 The formation of the complexes requires extrinsic and intrinsic events fine-tuned by extracellular inputs. cIAP (inhibitor of apoptosis)/BIRC (baculoviral IAP repeat-containing proteins) are closely linked to decision-making events. In this scenario, BIRC3, an E3 ligase,2, 3 ubiquitinates receptor interacting protein 1 (RIP1) and enables its interaction with the TAK1/TAB2/3 (TGFβ-activated kinase 1/(TAK1)-binding protein 2/3) complex, which in turn activates I kappa B kinase (IKK), and thus NF-κB. Alternatively, de-ubiquitinated RIP1 is engaged in tumor necrosis factor alpha (TNF-α)-triggered caspase 8-independent apoptosis.4 In these conditions, RIP3 has the ability to bind and phosphorylate RIP1 and is involved in apoptosis.5 While apoptosis is the predominant form of cell death in mild neuronal damage induced by ischemia-reperfusion in the penumbra region, necrosis is prevalent in the damaged core.6 Necroptosis, a form of programed necrosis, also involves RIP1. Necrostatin 1 (Nec1), a potent inhibitor of RIP1, prevents neuronal damage by reducing plasma membrane permeability and autophagy, and by improving functional impairment in a retinal ischemia-reperfusion model.7 In this manner BIRC3, deters TNF-α signaling from death-inducing signaling, thus preventing apoptosis and necroptosis.8
Docosahexaenoic acid (DHA; 22 : 6, n-3) is richly endowed and conserved in the retina and the brain.9 Upon uncompensated oxidative stress (OS), the onset of neurodegenerative diseases or ischemia-reperfusion, DHA is released by phospholipase A2 (PLA2) and stereoselectively oxygenated in carbons 10 and 17 by 15-lipoxygenase-1 (15-LOX-1).10 Neuroprotectin D1 (NPD1; 10 R,17Sdihydroxy-docosa-4Z,7Z,11E,15E,19Z hexaenoic acid) is released from the cell10 and activates a high-affinity binding site,11 setting in motion signaling to sustain homeostasis toward cell survival. NPD1 upregulates Bcl-2, Bcl-xl and Bfl-1/A1 and attenuates the expression of Bax, Bad and Bid.12, 13 Furthermore, NPD1 reduces caspase 3 activation induced by oxidative and proteotoxic stress.14, 15
To define the mechanisms by which NPD1 modulates neural cell survival, we used human retinal pigment epithelial (RPE) cells, which are neuroectoderm-derived post-mitotic cells of the retina, an integral part of the central nervous system.16, 17, 18 We report here that, in RPE cells, NPD1 induces cREL transcription and nuclear translocation that, in turn, mediates BIRC3 transcription and cell survival against OS. Furthermore, our data reveal that upregulation of NPD1 biosynthesis supports selective neuronal cREL and BIRC3 abundance, providing remarkable neurological recovery in experimental ischemic stroke.
Results
NPD1 stereoselectively enhances BIRC3 expression upon OS
NPD1 modulates the expression of proteins that counteract apoptosis, and it regulates inflammation under uncompensated OS.12, 18, 19 BIRC3 mRNA was upregulated by NPD1 after 6 h of OS treatment in 15-LOX-1 silenced cells, as demonstrated by microarray assay (Supplementary Figure S1a,Supplementary Table S1). In 15-LOX-1-silenced cells, which exhibit 90% decrease in NPD1 synthesis,10 BIRC3 mRNA levels are close to those observed in controls, suggesting that no BIRC3 induction occurs in the absence of NPD1. BIRC3 protein expression followed the same pattern of increment 2 h later (Supplementary Figure S1b,Supplementary Table S1). OS alone triggered BIRC3 expression in normal cells (Supplementary Table S1), but the addition of NPD1 potentiated this effect (Figure 1a,Supplementary Table S1), suggesting that endogenously synthesized NPD1 induced BIRC3 expression. To assess whether NPD1 specifically targeted BIRC3 transcription, and to determine the timeframe of activation, a qPCR assay was run that tested BIRC1 through BIRC8 on ARPE-19 cells after 2, 4 and 6 h of OS treatment in the presence or absence of 100 nM of NPD1 (Figure 1a and Supplementary Table S1). Only BIRC2, 3 and 4 showed at least a two-fold increase in expression under these conditions. From this subset, only BIRC3 and BIRC4 expression was increased differentially by NPD1. BIRC3 activation was observed as early as 2 h and peaked at 4 h. OS was an early activator of BIRC3 expression, but the effect declined after 6 h. The addition of NPD1, however, induced longer-lasting BIRC3 expression, and its decrease was not as steep as the expression in the absence of NPD1 (Figure 1a), suggesting that endogenous NPD1 may be responsible for the sustained activation observed. To evaluate the specificity of NPD1 on BIRC3 transcription activation, cells were treated with 100 nM functionally- and structurally-related lipids, including lipoxin A4 (LXA4) and maresin 1 (Figures 1bii–vi), and challenged with 600 μM H2O2/10 ng/ml TNF-α. LXA4 is a resolution-of-inflammation mediator derived from arachidonic acid,20 and maresin 1 is a DHA-derived mediator made by macrophages that also displays protective bioactivity.21 The data show that the NPD1 stereoisomer S-R (Figure 1bi), but not S-S or R-R (Figures 1biii and v), increased the expression of BIRC3 above the levels of OS-treated cells, indicating stereospecificity in the response (Figure 1b). On the other hand, neither maresin 1 nor LXA4 augmented BIRC3 abundance over the limits of OS, indicating that BIRC3 is a specific target of NPD1. To evaluate the NPD1-dependent increase in BIRC3, its messenger was transiently knocked down and its expression was triggered by OS and NPD1. Increasing siRNA concentrations lowered BIRC3 mRNA expression correspondingly, which reached a maximum at 20–50 pmol/ml of cultured medium (Figure 1c). Altogether, these results suggest that transcription of BIRC3 mRNA is stereospecifically and selectively enhanced by NPD1.
Stereospecificity of NPD1 bioactivity selectively upregulates BIRC3 expression. (a) BIRC1 to 8 in response to 600 μM H2O2/10 ng/ml TNF-α in the presence or absence of 100 nM NPD1 at 2, 4 and 6 h of treatment. (b) BIRC3 mRNA expression in response to 100 nM maresin 1, lipoxin-A4 and RvE1 along with NPD1 and its stereoisomers SS-NPD1 and RR-NPD1 and (c) in an siRNA dose-dependent curve. ARPE-19 cells were transfected with 0, 5, 10, 20, 50 and 100 pmol of siRNA per ml of culture media and treated with 400 μM H2O2/10 ng/ml TNF-α in the presence or absence of 100 nM NPD1. (i–vi) Structure of the lipid mediators used in (b). The bars represent the mean of three triplicates ± standard error. *P<0.05, NS=non-significant P-value. NPD1-treated samples=blue bars; OS+NPD1=light blue bars
TNFR1 signaling opposes NPD1-mediated BIRC3 transcriptional induction
BIRC3 interacts with tumor necrosis factor receptor-associated factor (TRAF) proteins associated with tumor necrosis factor receptor 1 (TNFR1), CD40 and other receptors as part of the canonical and non-canonical activation of NF-κB.2 ARPE-19 cells (which stably expressed shRNA that target TNFR1, as observed here) showed a 50% reduction in receptors (Supplementary Figures S2a and b) and were resistant to H2O2/TNF-α-induced OS (Figures 2a and b). To induce cell death, TNF-α was added to complement H2O2 since RPE cells are highly resistant to peroxide alone.12 TNFR1-silenced cells exposed to 6 h of 600 μM H2O2/10 ng/ml TNF-α in the presence or absence of 100 nM NPD1 showed higher levels of BIRC3 expression compared with control cells (Figure 2c), suggesting that TNFR1-dependent pathway competes in modulating BIRC3 transcription. To assess whether or not NPD1 bioactivity interferes with the canonical activation of NF-κB to promote the increase of BIRC3 expression, two steps of the pathway were checked. At an early point during NF-κB activation, the phosphorylation status of IκB (nuclear factor of kappa light polypeptide gene enhancer in B-cell inhibitor, alpha) was evaluated by western blot. Canonical activation of NF-κB requires phosphorylation and degradation of IκB by the proteasome. Under these conditions, the translocation of p65 dimers occurs. To test whether or not the TNFR1 pathway was impaired by NPD1, a time course for IκBα and β was performed. The phosphorylation of IκBα and IκBβ was not reduced by NPD1, and moreover, a decrease in total IκBα and IκBβ was noticeable at 30 min (Figures 2d and e) and 2 h when compared with resting cells (Supplementary Figures S3a and b). This suggests that NPD1 does not block the early canonical NF-κB activation pathway. Furthermore, after 30 min of treatment, NPD1 enhanced the phosphorylation of IκBα and IκBβ (Figures 2d and e), probably via a pathway other than the one activated by OS.22 Later in the pathway, transcriptionally-active NF-κB dimers were evaluated using a construct containing three p65/RelA and p50 high-scoring sites in tandem (Supplementary Table S5), driving the expression of the luciferase reporter gene. Accordingly, downstream of the TNFR1 pathway, at the level of NF-κB transcriptional activity, TNFR1-silenced cells showed half the activity of the negative control shRNA (NC) when treated with OS plus TNF-α (Figure 2f). For NPD1 and NPD1 treated cells, the same trend was also observed at 4 h, except OS did not alter luciferase expression (Supplementary Figure S2c). Although, the main NF-κB activity observed in control cells may be attributed to p65 dimers, the NF-κB binding sites contained in the construct showed lower scores for cREL (Supplementary Table S5). Thus, the increase in NF-κB transcriptional activity displayed by OS plus NPD1 in TNFR1-silenced cells may also be due to the activation of cREL dimers. These results suggest that the effect of NPD1 on NF-κB activation is TNFR1 independent. Also, we observed that NPD1 did not interfere with the early or late canonical activation of NF-κB, suggesting that NPD1 may enhance BIRC3 transcription by other means than blocking of TNFR1 signaling. Therefore, the opposing effects displayed by TNFR1-dependent activation and NPD1 on BIRC3 expression are not likely due to interaction/s with the two main pathways of NF-κB activation.
TNFR1 stably-silenced cells display enhanced BIRC3 expression and cell survival upon oxidative stress (OS). (a and b) TNFR1 and NC shRNA-expressing ARPE-19 cells were subjected to OS in the presence or absence of NPD1. (a) Representative pictures and (b) quantification of apoptotic TNFR1 and NC shRNA-expressing cells in the presence or absence of 50 nM NPD1. (c) BIRC3 expression induced by NPD1 upon OS by the means of real-time PCR in TNFR1-deficient cells. (d and e) Western blot showing the time-dependent phosphorylation of (d) IkBα and (e) IkBβ after 0, 15, 30 and 60 min of OS treatment in the presence or absence of 100 nM NPD1. (f) NPD1 effects on canonical NF-κB activation measured by the means of luciferase reporter assay in OS conditions. OS: 600 μM H2O2/10 ng/ml TNF-α. NPD1: 100 nM. Bars represent the mean of triplicates ± standard error of the mean. *P<0.05, NS=non-significant P-value. NPD1 treated samples=blue bars; OS+NPD1=light blue bars
cREL is the intracellular messenger of NPD1-mediated BIRC3 expression
Although it was previously shown that NF-κB is an enhancer of BIRC3 transcription,23 the identity of the dimer and the role of NPD1 signaling are currently unknown. In silico analysis of the BIRC3 promoter showed that of the three known NF-κB binding sites, located at 210, 197 and 147 bp upstream of the transcription start site (Supplementary Figure S4a), only cREL obtained the maximum score possible at the −210 site, while the other two sites (−197 and −147) either had a lower score or did not show any similarities (Supplementary Table S2). To determine the role of NPD1 in the activation of the BIRC3 promoter, NPD1 was added to ARPE-19 cells transfected with an array of promoter fragments (527, 247, 200, 174 and 93 base pairs upstream of the BIRC3 transcription start site, as shown in Figure 3a) driving the expression of luciferase in OS conditions. The effect of NPD1 on the construct −247 followed the same pattern as the BIRC3 mRNA (Figures 3b and c) at the 4- and 6-h time points; in addition, luciferase levels declined after 12 h (Supplementary Figure S4b). Then, the effect of NPD1 was evaluated in constructs containing disruptive mutations on the three binding sites (Figure 3a). Mutation of the −210 and −147 sites separately decreased the activity of the promoter, mainly the one triggered by OS alone (Figure 3c). Specifically, the disruption of the −147 site abolished NPD1 differential activation, suggesting that this latter binding site is the one influenced by NPD1 (Figure 3c). Simultaneous mutation of both sites completely abrogated the expression of the reporter gene (Figure 3c). The mkB2 construct displayed no differences in activity for the wild-type construct (−247), suggesting that the −197 site is not involved in regulation. In addition, the −197 site was previously reported to be silent for NF-κB.23 These results suggest that the −210 is an active binding site for cREL and that the −147 element has an important part in NPD1-mediated BIRC3 gene regulation (Figure 3d).
NPD1-mediated BIRC3 promoter activation. BIRC3 promoter was analyzed using a luciferase reporter assay. (a) Schematic representation of the constructs used for deletion and mutation. (b) BIRC3 promoter deletion analysis using constructs containing 527, 247, 200, 174 and 93 bp (depicted in a) upstream of the transcription start site. Transfected cells were treated with 130 μM H2O2/10 ng/ml TNF-α in the presence or absence of 200 nM NPD1. (c) Site directed mutation analysis on the NF-κB sites. Cells were treated with 400 μM H2O2/10 ng/ml TNF-α in the presence or absence of 200 nM NPD1. (d) Luciferase activity was standardized by GFP fluorescence. Bars represent the mean of triplicates ± standard error of the mean. *P<0.05, NS=non-significant P-value. NPD1-treated samples=blue bars; OS+NPD1=light blue bars
NPD1 triggers cREL gene transcription and nuclear translocation
To define the role of cREL in the activation of BIRC3 transcription, we focused on changes in the distribution of cREL in response to OS, and on how NPD1 affects this process. When cells were incubated with H2O2 and TNF-α to induce OS, an increase in nuclear signaling was observed at 2 h, followed by a steep decrease at 4 h, and then another rise at 6 h (Figures 4a and b). At 6 h, no difference was observed between OS and OS plus NPD1 treatments, probably because of the ongoing endogenous synthesis of NPD1. Nuclear translocation of cREL was confirmed by western blot of nuclear extracts obtained after 2 h of treatment (Figure 4d). The increase in cytoplasmic cREL-positive cells mirrored that observed in the nuclei at the 2-h time point (Figures 4b and c). On the contrary, at later time points cytoplasmic-positive cells did not vary with treatments, suggesting that the 2-h wave of translocation leads to de novo cREL expression (Figure 4c). Furthermore, the total amount of cREL protein increased with the addition of NPD1 at 4 h of treatment (Figure 4e). ChIP assay confirmed enhanced activity, showing that cREL binding to genomic DNA was higher in the presence of NPD1 than in controls and cells treated with OS alone (Figure 4f). As described above, although its value was lower than cREL, p65/RelA scored significantly positive for the sites in the BIRC3 promoter (Supplementary Table S2); thus, p65/RelA may be a second activator of BIRC3, perhaps as part of the p65/RelA-cREL dimer (Figure 3d). Because of this, p65/RelA was co-immunoprecipitated with cREL at 4 h of treatment as well (Figure 4g). Remarkably, the NF-κB complexes were enriched in cREL when cells were treated with OS plus NPD1 and p65/RelA levels were down (Figure 4g). These observations support the NPD1-triggered rise in cREL expression that took over the NF-κB-active dimer. To examine whether or not cREL is part of the signal amplification loop, the expression of cREL was assessed in cells exposed to OS in the presence and absence of NPD1 at 2, 4 and 6 h. cREL mRNA increased two-fold at 2 h and almost five-fold at 4h, and at this latter time the presence of NPD1 increased expression to levels greater than those attained by OS alone (Figure 4h).
cREL nuclear translocation and binding to BIRC3 promoter to induce the activation of its expression. Changes of distribution, activity and expression of cRel were assessed at three time points (2, 4 and 6 h) in ARPE-19 cells treated with 400 μM H2O2/10 ng/ml TNF-α in the presence or absence of 100 nM NPD1. (a–c) Immunocytochemistry of cREL in cells: (a) representative pictures showing the distribution of the cREL signal (red). Nuclei were stained with DAPI (blue). (b and c) Portion of cells depicting cREL nuclear or cytoplasmic signal. (d and e) cREL protein content evaluated by western blot (d) in the nuclear fraction standardized using TBP at 2 h and (e) in whole cells standardized by GAPDH after 4 h of OS or OS+NPD1. (f) ChiP assay at 4 h showing the binding of cREL to BIRC3 promoter. The co-immunoprecipitated genomic DNA was amplified and standardized by the input. (g) Co-immunoprecipitation of cREL and p65/RelA at 4 h of treatment standardized by GAPDH. (h) p65/RelA, RelB and cRel expression determined by real-time PCR. (i–k) Silencing of cRel induced changes in the expression of: (i) cREL, (j) RelB and (k) BIRC3 in human RPE (hRPE) cells established by the means of real-time PCR. Concentrations of 2.5, 10 and 50 pmol siRNA per ml of cell culture medium were used to show a concentration-dependent effect on the expression at 4 h. (l and m) Schematization of the temporal pattern of (l) cREL, RelB and BIRC3 expression and (m) cREL translocation. The values are represented as the mean of triplicates ± the standard error. *P<0.05, NS=non-significant P-value. NPD1-treated samples=blue bars; OS+NPD1=light blue bars
Intriguingly, RelB expression also showed an elevation at 4 h. After 6 h, cREL activation was lessened, but RelB levels were still higher than controls (Figure 4h), suggesting a different time pattern. RelA showed no differences between treatments or times (Figure 4h). In silico analysis of cREL and RelB promoters showed that both promoters contain cREL binding sites (Supplementary Tables S3 and S4).
To confirm whether cREL is a key mediator of BIRC3 activation, we silenced its expression in human RPE cell primary cultures. cREL expression was decreased in a dose-dependent manner in human RPE primary cells (Figure 4i), providing evidence for NPD1-induced cREL transcription; RelB expression also was affected (Figure 4j). cREL silencing decreased the levels of NPD1-dependent RelB expression to those achieved by OS alone, suggesting there are at least two components involved in RelB activation: one triggered by NPD1 and the other by OS (both of which are mediated by cREL in a different manner and probably by different dimers). This is consistent with the model proposed for the BIRC3 promoter in Figure 3d. Finally, BIRC3 mRNA was decreased by silencing cREL in the presence and absence of NPD1 (Figure 4k); thus, BIRC3 mRNA induction depends on cREL synthesis. The temporal expression pattern followed by these three genes is illustrated in Figure 4l, and the translocation of cREL is shown in Figure 4m. Altogether, these results suggest that NPD1 triggers cREL mobilization into the nucleus in a periodic manner to jumpstart the expression of a cluster of pro-survival genes, likely as a consequence of increased abundance of cREL homodimers.
NPD1 fails to rescue BIRC3-silenced RPE cells from OS-induced apoptosis
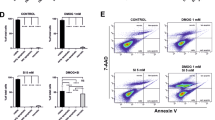
To assess the significance of BIRC3 in NPD1-triggered survival signaling, BIRC3 or cREL was silenced and apoptosis was induced by OS. NPD1 failed to rescue BIRC3- and cREL-silenced ARPE-19 and human RPE cells (Supplementary Figures S5 and S6) from apoptosis (Figures 5a–e). In addition, the overexpression of both cREL and BIRC3 (Figures 5f and g; Supplementary Figure S7) decreased OS-induced cell death to the levels observed with NPD1 treatment (Figures 5f–h). Furthermore, overexpression of cREL led to an increase in the BIRC3 protein (Figure 5f). In addition, the overexpression of cREL and BIRC3 siRNA-resistant open reading frames rescued the decreased expression and increased apoptosis induced by OS in the corresponding silenced cells (Figures 5i and j). The amount of transfection reagent required to perform the co-transfection induced a sensitization of the cells, lessening the differences between treatments and reducing the effects of overexpression, as shown in Figure 5h. Previous reports show that NPD1 prevents caspase 3-dependent apoptosis.12, 14, 18 To determine whether the level of active caspases 3 and 7 regulated by NPD1 is mediated by BIRC3, silenced cells were challenged with OS. Fifty percent of the BIRC3 knockdown cells were caspase positive compared with 35% of the control cells, and NPD1 reduced this percentage to control levels only in cells expressing normal levels of BIRC3 (Figures 5k and l). Also, Z-VAD, a pan caspase inhibitor, consistently prevented activation of caspases 3 and 7. These results suggest that the NPD1-mediated increase in BIRC3 levels thwarts activation of effector caspases 3 and 7. It has been proposed that BIRC3 has the capability of binding effector caspases, but that it is not able to inhibit their activity.24 To investigate the mechanism by which NPD1/BIRC3 induces a decrease in activated effector caspases, and whether or not RIP1 is involved in NPD1/BIRC3-mediated survival, Nec1, an inhibitor of RIP1, was included in the treatment. BIRC3 knockdown prevented NPD1 from affecting OS-induced apoptosis. Nec1 reversed this effect, showing that RIP1 is involved in the apoptotic process prevented by NPD1 (Figure 5m). To test the role of NPD1-induced BIRC3 in the prevention of necrosis, we silenced BIRC3 and applied OS, Z-VAD and/or Nec1. OS alone increased the percentage of necrotic cells, and NPD1 attenuated these effects (Figure 5n). When BIRC3 was silenced, as expected, the levels of necrosis were not reduced by NPD1. The addition of Nec1, or both Nec1 and NPD1 together, to the treated cells decreased the levels of necrosis and apoptosis to control levels. These results suggest that RIP1 is involved in the necrosis and apoptosis triggered by OS and that its relative abundance may be controlled by NPD1-dependent BIRC3. Previously, it was shown that some growth factors have the ability to induce NPD1 synthesis and promote cell survival.13 To assess whether these two survival-promoting effects were achieved through a common pathway, we silenced cREL and challenged the cells with OS in the presence or absence of DHA plus the growth factors PEDF, FGF, CNTF or BDNF. cREL silencing exacerbated the apoptosis induced by OS in PEDF-, CNTF- and FGF-treated cells, following the same trend observed in NPD1-treated cells (Figure 5o, lower panel). On the other hand, no change was observed for BDNF silenced and control cells; since both types were protected equally, this suggest that its effect is cREL independent. It was shown previously that these four growth factors induce NPD1 synthesis in RPE cells13 and that endogenous NPD1 synthesis is noticeable 4–5h after treatment.10 To test whether cREL was activated in direct response to the addition of these growth factors or through NPD1 signaling, the translocation of overexpressed cREL fused to green fluorescent protein (GFP) was assessed after 2 h of treatment. None of the growth factors induced translocation of cREL above the control level (Figure 5o). As expected, exogenously-added NPD1 increased cREL translocation at 2 h, consistent with the results mentioned above (Figures 4b and d). When cREL was silenced, the protection exerted by the growth factors PEDF, FGF and CNTF, when added along with DHA, was abolished following the same trend as NPD1 (Figure 5o). This provides evidence that endogenous production of NPD1, when triggered by these growth factors, activates cREL-dependent survival against OS-induced apoptosis. Overall, these results suggest that exogenously-added and endogenously-synthesized NPD1 exerts its protection through the increase in cREL-mediated BIRC3 expression. BIRC3 instead interacts with the RIP1 complexes, preventing effector caspases 3 and 7 from being activated, and thus decreasing apoptosis.
NPD1 fails to rescue BIRC3 silenced cells. (a–e) Apoptosis noted as percentage of Hoechst- or TUNEL-positive cells was assessed on ARPE-19 (a–c) or primary human RPE (hRPE) cells (d and e). (a) Representative images of TUNEL staining performed on ARPE-19 cells, (b and c) Quantification of TUNEL and Hoechst-positive cells when transfected with BIRC3 or control siRNA and treated with 0, 400 and 600 μM H2O2/10 ng/ml TNF-α with or without 200 nM NPD1. BIRC3 (d), cREL (e) or control siRNA-transfected hRPE cells treated with 600 μM H2O2/10 ng/ml TNF-α in the presence or absence of NPD1 100 nM. cREL and BIRC3 (f) and BIRC3 (g) protein content when cREL and BIRC3, respectively, were overexpressed. (h) Percentage of apoptosis in ARPE-19 cells overexpressing cREL or BIRC3 when confronted with OS in the presence or absence of NPD1. (i and j) Overexpression of (i) cREL or (j) BIRC3 using a wild-type open reading frame (ORF) or one carrying silent mutations at the siRNAs binding sites (ORFmut) to prevent its silencing and rescue from the knocked down phenotype. Upper panels show representative western blots for the corresponding protein content in each sample. Lower panels depict percentage of apoptosis for each treatment. (k and l) Activation of effector caspases 3 and 7 as a result of OS treatment in BIRC3-silenced cells. (k) Representative nuclei and staining of cells: upper panel, control; lower panel, OS. (m and n) Apoptosis and necrosis measured by the means of AnnexinV and 7-Amino actinomycinD when cells were treated with OS and NPD1 in the presence of z-VAD, a pan caspase inhibitor or Necrostatin 1 (Nec1). (o) cREL translocation (upper panel) and percentage of apoptosis of control and cREL-silenced (lower panel) ARPE-19 cells subjected to OS with the addition NPD1 or DHA plus 10 ng/ml of PEDF, FGF, CNTF or BDNF to endogenously induce NPD1 synthesis. (p) Schematization of the model obtained by the integration of the data obtained in this report and the context. Bars represent the mean of triplicates+standard error of the mean. *P<0.05 NS=non-significant P-value. NPD1-treated samples=blue bars; OS+NPD1=light blue bars; DHA+growth factors=teal bars; OS+DHA+growth factors=light teal bars
NPD1 biosynthesis after systemic DHA in ischemic stroke selectively increased neuronal cREL and BIRC3 abundance and neurological recovery
To test the significance of NPD1 as a modulator of BIRC3 expression in vivo, we used experimental ischemic stroke by means of middle cerebral artery occlusion (MCAo) (Figures 6a–c). Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS)-based mediator lipidomic analysis revealed that upon DHA treatment, 17-HDHA (the stable derivative of HpDHA, the precursor of NPD1) as well as NPD1 were increased greatly only 1 day after the ischemic event on the ipsilateral side (Figure 6e). Supplementary Figure S8 shows the mass spectrometry fragmentation pattern of these lipids isolated from the ipsilateral side of the brain. Rats treated with DHA consistently exhibited an improved composite neurologic score compared with the saline-treated group on days 1, 3 and 7 after stroke (Figure 6d). These rats displayed improved tactile placement (proprioceptive, lateral and dorsal), forelimb placement, and an overall increased neurological score (Figure 6d).
BIRC3 mediates the pro-survival response induced by the DHA/NPD1 pathway in an ischemia-reperfusion stroke model. (a–c) Model for inducing ischemia-reperfusion by middle cerebral artery occlusion (MCAo) in rats. (a) Timeline showing surgery, treatment and tests performed. (b) Coronal brain diagram (bregma level –0.3 mm) showing locations of regions for western blot and immunohistochemistry for Figure 7, and lipidomic analysis (A: anterior; P: posterior). (c) Diagram of MCAo model obtained by introducing intraluminal filament (red). (d) Total neurological score (normal score =0, maximal deficit=12), tactile placing (proprioceptive, lateral, dorsal reactions; normal score=0, maximal deficit=2) in rats after MCAo. DHA treatment improved the total and placing deficits on days 1, 3 and 7 compared with the saline-treated group. (e) Content of NPD1 and a second product of the stabilized precursor, 17HDHA, in penumbra regions of rats subjected to MCAo and treated with DHA or vehicle as a control. Data are means±standard error of the mean; n=6 per group. *P<0.05 in repeated-measures, ANOVA followed by Bonferroni test. DHA treatment=teal bars
BIRC3 abundance also increased in the ipsilateral side of the brain of DHA-treated animals on day 1, but not on days 3 or 7, after MCAo (Figure 7a,Supplementary Figure S9). NPD1-dependent BIRC3 co-localized with NeuN, but not with GFAP (Figures 7f and h and Supplementary Figure S10). Quantification of neuronal BIRC3 showed an increase in BIRC3 abundance (Figure 7h). Moreover, the overall content of cREL was increased by DHA treatment (Figures 7b and d), as was the nuclear translocation (Figures 7c and e). These results demonstrate that DHA, when administered systemically after MCAo, leads to NPD1 synthesis, enhanced ipsilateral neuronal cREL translocation, and BIRC3 expression. Thus, these studies reveal that the neurological recovery may result from the endogenous NPD1 synthesis and its consequent neuronal-specific cREL-mediated activation of a cluster of pro-survival genes, including BIRC3, following experimental stroke as a new pro-survival pathway.
DHA/NPD1 induce translocation of cREL and increased BIRC3 in vivo. (a) Western blot on days 1, 3 and 7 after DHA treatment performed on the posterior section. (b–e) Translocation of cREL in neurons of the penumbra, areas A1-3 and P1-3 (see Figure 6b). (b) Representative images of nuclear translocation of cREL in saline- and DHA-treated animals in the P2 region 1 day after treatment. (NeuN red; c-REL green). (c) High magnification. cREL (d) total (upper panels) and (e) nuclear (lower panels) quantification from areas A1 to 3 (upper panels) and P1-3 (lower panels). (f) BIRC3 (red) and NeuN (green), and (g) BIRC3 (red) and GFAP (green) double staining on day 1 after stroke. (h) Quantification of the co-localization studies in (f and g). Data are means±S.E.M.; n=6 per group. *P<0.05 in repeated-measures, ANOVA followed by Bonferroni test. DHA treatment=teal bars
Discussion
For the first time, evidence is provided that NPD1 activates pro-survival transcription factor cREL in cells in culture as well as selectively in neurons in an in vivo model of ischemic stroke. Moreover, consequent neuronal-specific BIR3 expression takes place. NF-κB dimers are shuttled from the cytoplasm into the nucleus either by phosphorylation, ubiquitination and degradation of inhibitors of NF-κB (IκBs) or, in the case of p100/p50 and p105/p52, by cleavage of the inhibitory domain.25 The mechanisms by which dimers of cREL are activated remain largely unknown. Here, we uncover that NPD1 signaling induces an increased translocation of cREL dimers into the nucleus (Figures 4b, 5o, 7b, c and e). Like other components of the NF-κB system, cREL presents a periodic behavior.26, 27, 28, 29 This distinctive feature was noticed in the nuclear translocation of NF-κB (Figure 4b), where the 2-h time point showed nuclear signaling in a high proportion of the cells, while the 4-h time point displayed very low levels of translocation in accordance with the time span between periods (where the periodic frequency is around 3 h and 45 min).27 In this context, our data point to a first wave of nuclear translocation (probably where the RelA/cREL dimer dominates), in which the initial transcriptional amplification of cREL begins (Figures 3d, 4b–d, 5o and p), followed by a second wave where the predominant dimer is cREL-cREL, which renders a sustained transcription of BIRC3 (Figures 1a, 3d, 4c, e, h and 5p). Thus, the role of NPD1 may be to alter the amplitude and period of the NF-κB cyclic response, and by doing so accelerate and heighten the pro-cell survival response (Figures 4l and m).
The role of BIRC3 in caspase inactivation is also unclear. BIRC3 was proposed to bind but not inhibit caspases 3 and 7, unlike their close family member XIAP.24 However, here we show that BIRC3-silenced cells displayed a higher content of activated caspases 3 and 7 than control cells when treated with OS (Figures 5k and l). Furthermore, the silencing of BIRC3 (Figures 5a–d and l–n) abolished the effects of NPD1. This strongly suggests that NPD1 acts through BIRC3 to decrease caspase activity. BIRC3 is involved in the steps that precede the switch between NF-κB activation, apoptosis and necrosis by regulating the formation of complexes I, II and III, especially the formation of complex II, which requires activation of RIP1, a ubiquitination target of BIRC3.30 Complex II, or death-inducing signaling complex (DISC), is one of the initiators of effector caspases 3 and 7 along with the apoptosome.1 Thus, BIRC3 may act through RIP1 on the caspase 8 complex to prevent the conversion of pro-caspase 3 into its active state. Accordingly, overall levels of apoptosis were decreased by the addition of Nec1 (Figure 5m). BIRC3 silencing also increased necrosis (Figure 5n), as described earlier, when SMAC mimetics were added.31 The necrosis observed after BIRC3 knockdown was RIP1 dependent, since the process was abolished by the addition of Nec1 (Figure 5n). The stereospecificity of NPD1 bioactivity (only to the 17S-10R and not to the R-R or S-S stereoisomers) implies that specific signaling takes place (Figure 1b). LXA4 and resolvin E1 (RvE1), which also are engaged in cytoprotection but are not structurally-related lipids to NPD1, are ineffective in inducing BIRC3 expression. Upon OS, PLA2 releases DHA, and 15-LOX-1 catalyzes NPD1 synthesis.10 Ischemic stroke involves a complex multiphasic cascade of cellular and molecular events, with an early ischemic phase followed by a secondary progression of injury from the neuroinflammatory responses. Initially, toxic nitric oxide, reactive oxygen species, proteases, lipases, phospholipases, cytokines and chemokines recruit peripheral macrophages, neutrophils, dendritic cells and later T and B cells.32
Overall, neuroinflammation, excitotoxicity and enhanced OS threaten the integrity of neurons and other neural cells. DHA improves recovery after ischemic stroke33, 34 and acts, through its conversion into NPD1, to halt homeostasis disruptions and cell death signals.9 Thus increased NPD1 synthesis (Figure 6e), as a consequence of DHA administration in a stroke MCAo model, induces selective neuronal cREL translocation followed by BIRC3 expression (Figures 7a, f and h), resulting in remarkable neurological recovery. Thus, as was the case in our cellular model, cREL was translocated into the nucleus to a greater extent in DHA-treated animals than in saline-treated ones (Figures 7c and e), suggesting that NPD1 produced by the conversion of systemically-administered DHA (Figure 6e) acts through c-REL-mediated BIRC3 transcriptional activation to elicit its neuroprotective mechanism. The cREL neuroprotective effects observed here are in agreement with previous work by Pizzi’s group on brain ischemia,35, 36 which poses that cREL- and p65/RelA-containing dimers are balanced as the main factors in neuronal survival. In our cellular model, we also found that when the cREL protein increases, it leads to survival and a decrease in p65/RelA, in response to NPD1 (Figures 4d, e, g and 5f–h and Supplementary Figure S7a). These findings will help to further unravel the endogenous signaling that sustains retinal and brain cellular integrity, thus providing a greater understanding of these mechanisms that could lead to novel therapeutic approaches for neuroprotection.
Materials and Methods
Reagents and constructs
The primary antibodies to detect BIRC3, TNFR1, TBP, cREL, phospho-IκBα and IκBβ, and RelA for western blot and immunocytochemistry were obtained from LifeSpan Biosciences (Seattle, WA, USA), Abcam (Cambridge, MA, USA) and Cell Signaling (Danvers, MA, USA), respectively. Anti-GAPDH antibody was obtained from EMD Millipore (Billerica, MA, USA). Fluorescent-tagged negative siRNA was acquired from Qiagen (Venlo, The Netherlands), secondary antibodies for immunocytochemistry were purchased from Molecular Probes (Life Sciences, Carlsbad, CA, USA), and antibodies used for western blot were obtained from GE Healthcare (Piscataway, NJ, USA). Buffers and supplies for western blot and cell cultures were obtained from Invitrogen (Carlsbad, CA, USA), Bio-Rad (Hercules, CA, USA) and Sigma (Saint Louis, MO, USA), and fetal bovine serum (FBS) was from Tissue Culture Biologicals, Inc. (Seal Beach, CA, USA). BIRC3/c-IAP2 promoter-driven luciferase reporter constructs, along with the mkB1, mkB2, mkB3 and mkB1,3 mutants, were generously provided by Tae H Lee (Yonsei University, Seoul, South Korea). BIRC3 and cREL expression was silenced using a mixture of three siRNA commercially available as Trilencer-27 siRNA knockdown duplexes kit (Origene, Rockville, MD, USA). Wild-type BIRC3-, cREL- and cREL-fused-with-GFP-expression constructs were obtained from Origene. BIRC3 and cREL siRNA-resistant expression constructs were designed by including silent mutations in the siRNA binding sites and synthesized by GeneArt (Life Technologies-Thermo Fisher, Carlsbad, CA, USA). siRNA-resistant BIRC3 and cREL ORF were designed by JMC and synthesized by GeneArt (Life Technologies, Carlsbad, CA, USA). The mutated ORFs were subcloned in pCMV-XL5 (Origene) using SacI and XbaI restriction sites. Z-VAD was obtained from R&D Systems (Minneapolis, NE, USA), and Nec1 was purchased from Enzo Life Sciences (Farmingdale, NY, USA). Active growth factors FGF, CNTF, PEDF and BDNF were obtained from Sigma.
Computational analysis
To analyze the consensus binding sequences of the BIRC3, cREL, RelB and RelA promoters, three searching engines were used: (1) TRED (http://rulai.cshl.edu/TRED), which uses the JASPAR database, (2) TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html) and (3) TFBind (http://tfbind.hgc.jp/); the latter two engines use the TRASFAC database.37, 38, 39 The validated BIRC3, cREL, RelB and RelA promoter sequences were obtained from previous publications.22, 40 Transcription of the RelB gene was regulated by NF-κB.41, 42
Cell culture, nuclei extraction, transfection, treatments and luciferase assay
Silenced 15-LOX-1 cells were prepared as described previously.12 Primary human RPE cultures were obtained from human eye cups, provided by the National Disease Research Interchange (NDRI), as described previously.14 ARPE-19 cells and 15-LOX-1d cells were plated at a 125 000 cell per ml density and grown in DMEM/F12 containing 10% FBS and 1 × Penicillin/streptomycin at 37 °C 5% CO2, 99% relative humidity for 24–48 h until they reach confluency. The three cell types were serum-deprived for 8–16 h before the addition of NPD1 and its stereoisomers S-S and R-R, DHA/pigment epithelium-derived factor (PEDF), LXA4, maresin, RvE1 and H2O2/TNF-α. ARPE-19 cell nuclear extracts were prepared using a nuclear extraction kit (Millipore) following the manufacturer’s protocol. Briefly, 100% confluent ARPE-19 cells were serum-starved for 8 h and then treated with 600 μM H2O2/ 10 ng/ml TNF-α in the presence and absence of 400 nM NPD1 for 2 h. Cells were collected and processed in a cytoplasmic lysis buffer, passed through a 27 gauge syringe, and pelleted at 250 g. Pellets were re-suspended in nuclear lysis buffer, passed again through a 27 gauge syringe to break down the nuclei, kept on a rotator shaker for 60 min, and then centrifuged at 13 000 r.p.m. Supernatants were then used to assess the nuclear content of cREL by western blot.
For the luciferase assay, cells were co-transfected with plasmid containing the BIRC3 promoter constructs depicted in Figure 3a and described in detail previously,43 and GFP or β-galactosidase expression vectors were co-transfected using Lipofectamine 2000 (Life Technologies, Grand Island, NY, USA). For activation of canonical NF-κB, cells were transfected with plasmid containing p65/p50 promoter consensus sequence along with both positive (constitutively-active promoter) and negative controls. In all cases, transfection was performed 24 h after plating at an 80–95% confluency using Lipofectamine 2000 (Life Technologies) following protocols described previously.12 BIRC3 and cREL siRNAs were transfected in ARPE-19 and hRPE cells using Lipofectamine 2000 (Life Technologies) following the manufacturer’s recommendations. Transfection of the three siRNAs separately did not provide the same yield as a mix of three (Supplementary Figures S11 and S12). Ten pmol per ml of total medium of fluorescent-tagged non-specific siRNA (Qiagen, Valencia, CA, USA) was added to the mixture of three siRNAs to assess the transfection efficiency (TE) in each sample. Twenty-four hours after transfection, cells were treated as described above or fixed to determine TE. TE averaged 85%, and experiments with TE lower than 80% were discarded. To determine translocation of cREL, two approaches were used: immunocytochemistry of cells (experiments depicted in Figures 4a–c) or immunohistochemistry of tissue (experiments depicted in Figures 7b–e) treated or transfected with cREL ORF fused to GFP (Figure 5o, upper panel). The latter experiment was performed by transfecting ARPE-19 cells using Lipofectamine 2000 as described above. The translocation was assessed by counting the nuclear GFP signal as a percentage of the total cells (Hoechst nuclear staining).
To evaluate the effect of growth factors, cells were treated with 20 ng/ml FGF, CNTF, BDNF or PEDF plus DHA, or NPD1 alone simultaneously with 400 μM H2O2 and 10 ng/ml TNFα (OS). Overexpression of cREL and BIRC3 wild-type and siRNA-resistant ORFs was performed similarly by transfecting the ORF contained in the expression vector pCMV6-XL5, as described above, and treating the cell 24 h later. cREL and BIRC3 expression in silenced and overexpressing cells was tested by real-time PCR (Supplementary Figure S7) and western blot (representative blots in Supplementary Figures S5 and S6). To assess luciferase activity, cell lysates were obtained using Passive Lysis Buffer (Promega, Madison, WI, USA) and mixed with a luciferase assay reagent (Promega). Chemiluminescence produced by luciferase and fluorescence from GFP was detected using Appliscan 2.3. Data were analyzed using SkanIt 2.3 (Thermo Scientific, West Palm Beach, FL, USA). In all cases, whenever transfection was performed, the transfection efficiency was tested using GFP expression vector as described previously.14
TNFR1-stably silenced cells
Cells stably expressing the shRNA targeting TNFR1 were obtained from the ARPE-19 parental cell line using a protocol described previously.12 Briefly, FuGENE 6 (Roche, Applied Science, Brandford, CT, USA) was used to transfect cells using four different vectors, each one carrying sequences targeting TNFR1 and non-specific shRNA included in the SuRE silence system (SABiosciences, Qiagen). Stable transfectant/neomycin-resistant cells were selected by growing them in a DMEM/F12 medium containing 10% FBS and 0.5 mg/ml Geneticin (Invitrogen). Surviving cells formed colonies that were trypsinized separately, grown and then tested for TNFR1 content using western blot (Supplementary Figure S2a) and real-time PCR (Supplementary Figure S2b).
ChIP aasay, immunoprecipitation and western blot
The ChIP assay was performed as follows: ARPE-19 cells were grown in 10 mm culture dishes to confluency. After treatment, cells were washed with PBS 1 × and proteins were cross-linked to DNA by addition of 1% formaldehyde for 10 min followed by addition of 2.5 mM glycine for 5 min. Cells were scraped with ice-cold PBS 1 × and centrifuged at 400 × g. The pellet was washed three times with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1), and DNA was fragmented by sonication and incubated in ChIP dilution buffer (16.7 mM Tris-HCl, pH 8.1, 1.2 mM EDTA, 1.1% Triton-X, 167 mM NaCl, 0.01% SDS and protease inhibitors) with cREL antibody (Bethyl, Montgomery, TX, USA) at 4 °C overnight. Samples were incubated with protein A/G plus agarose beads for 3 h, centrifuged, washed and eluted with 1% SDS and 0.1 M NaHCO3. Protein/DNA complexes were treated with Proteinase K. DNA was purified using the phenol/chloroform extraction method and amplified by PCR using the primers depicted in Supplementary Table S6. For immunoblot analysis of the immunoprecipitated proteins, the eluted samples were treated with DNAse I (Invitrogen). Western blot was performed from lysates obtained using RIPA buffer supplemented with a protease inhibitor cocktail (Sigma). Total protein (30–50 mg) was mixed with Laemmli buffer containing DTT and loaded in Novex 4–12% precast gels and ran in an X-Cell running system at 120 V for 1.5 h. The transference was performed using the Trans Blot Turbo dry transferring system (Bio-Rad) on low fluorescent background PVDF membranes (GE Healthcare). Membranes were incubated with the corresponding primary antibodies overnight with fluorescent-tagged secondary antibodies for 1 h and then dried. Data were acquired using ImageQuant LAS4000 (GE Healthcare). Densitometric analysis was performed using ImageQuantTL software (GE Healthcare).
Microarray assay, PCR array and qPCR
Total RNA was isolated using an SV Total RNA isolation system (Promega) or RNeasy Plus Mini Kit (Qiagen). For qPCR array, cDNA was synthesized using an RT2 First Strand Kit RT2 Profiler PCR pathway focused on an apoptosis-related gene array (SABiosciences, Frederick, MD, USA). Expression screening experiments using Affymetrix GeneChip HG-U133 plus 2.0 (Santa Clara, CA, USA) were carried out by the Microarray Core at the Louisiana State University Health Sciences Center, New Orleans. ARPE-19 cells were treated as described above and total RNA was extracted, tagged and hybridized in the microarray. Data obtained were analyzed using one-way ANOVA and false positive test. qPCR to assess expression of BIRC family members, cREL, RelA, RelB and TNFR1 was performed using CFX384 iCycler (Bio-Rad). Briefly, 1 μg of total RNA was reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad), and the reaction was carried out with IQ TM SYBR Green Supermix (Bio-Rad); primers are depicted in Supplementary Table S7. Data were analyzed using the delta-delta Ct analysis method. Included data show a single unique dissociation peak in the melting curve.
Immunocytochemistry, Hoechst staining, TUNEL assay and caspase activity assay
Immunocytochemistry assays were performed in 8-well slide chambers, and Hoescht staining was performed in 6-well plates following a previously-described protocol.14 Briefly, cells were fixed in 4% paraformaldehyde for 20 min, permeabilized with Triton X-100 0.1% in PBS, and non-specific epitopes were blocked in 10% normal serum and 1% Bovine sero-albumin (BSA) in 1 × PBS for 1 h at room temperature. Immunostaining was performed by incubating primary antibody overnight at 4 °C. Samples were stained for 2 h at room temperature with Alexa Fluor 555 and 488 conjugated secondary antibodies (MeridianLife Science Inc., Memphis, TN, USA) and nuclei were stained with Hoechst or DAPI. Hoescht staining was carried out using 2 μM Hoechst33258. Pictures were taken with Nikon DIAPHOT 200 microscope. Images were then imported into the image analysis software ImageJ (rsb.info.nih.gov/ij/), and processed using custom macros. TUNEL staining was performed using DeadEnd fluorometric TUNEL system (Promega) following the manufacturer’s directions. Active caspases 3 and 7 were assayed using FAM-FLICA caspases 3 and 7 reagent, FAM-DEVD-FMK (Immunochemistry Technologies, Bloomington, MN, USA) following the manufacturer’s protocol. Detection of necrosis was carried out using a GFP-Certified Apoptosis/Necrosis Detection Kit (Enzo Life Sciences) following the manufacturer’s protocol. In all cases, slides were mounted in Dako Fluorescent Mounting Medium (Dako North America Inc., Carpinteria, CA, USA) and analyzed using an Axioplan 2 deconvolution microscope (Thornwood, NY, USA) and Zeiss LSM 510 Meta Confocal Laser Scanning microscope (Thornwood, NY, USA). Images were obtained and processed using SlideBook 4.2 and 5.0 software (Intelligent Imaging Innovations Inc., Denver, CO, USA) and LSM 510 Meta software (Thornwood, NY, USA).
Animals and focal cerebral ischemia model
All studies were approved by the Institutional Animal Care and Use Committees of the Louisiana State University Health Sciences Center (New Orleans, LA, USA) and Loma Linda University (Loma Linda, CA, USA). Male Sprague-Dawley rats (Charles River Lab., Wilmington, MA, USA) weighing 260 349g were fasted overnight but allowed free access to water. Anesthesia was induced with 3% isoflurane in a mixture of 70% nitrous oxide and 30% oxygen. All rats were orally intubated and mechanically ventilated. Temperature probes were inserted into the rectum and in the left temporalis muscles to maintain temperatures at 36–37 °C during surgical procedures. The right femoral artery and vein were catheterized for blood sampling for arterial gases, pH, and plasma glucose.
Middle cerebral artery occlusion
Rats underwent 2 h of right MCAo by an intraluminalfilament, as previously described.34 In brief, the right common carotid artery bifurcation was exposed through a midline neck incision and the occipital artery branches of the external carotid artery were isolated, ligated and dissected. After careful isolation of the internal carotid artery, a 3-0 monofilament coated with poly-L-lysine was advanced through the internal carotid artery to the MCA until mild resistance was felt. The neck incision was closed with a silk suture and the animals were then allowed to recover. After 2 h of MCAo, rats were reanesthetized with the same anesthetic combination and intraluminal sutures were carefully removed.
Behavioral tests
The animals underwent neurobehavioral assays by an observer blinded to the treatment groups at 60 min (during MCAo) and then on days 1, 3 and 7 after MCAo. The battery consisted of the postural reflex test and the forelimb placement test for visual, tactile and proprioceptive stimuli. Neurological function was graded on a scale of 0–12 (normal score=0; maximum score=12) as previously described.34
Treatment and experimental groups
Docosahexaenoic acid (DHA; 5 mg/kg, Cayman, Ann Arbor, MI, USA) or vehicle (0.9% saline) was administered intravenously into the femoral vein at a constant rate over 3 min using an infusion pump at 3 h after onset of MCAo. For western blot studies of DHA- or saline-treated rats, animals were killed on days 1, 3 and 7 (six groups, n=6 per group). For immunohistochemistry studies of DHA- or saline-treated rats, animals were killed on day 1 (two groups; n=7 per group).
Statistical analysis
Data are presented as mean values±S.E.M. Repeated measures analysis of variance (ANOVA) followed by Bonferroni procedures to correct for multiple comparisons was used for intergroup comparisons. Two-tailed Student’s t tests were used for two-group comparisons. Differences at P<0.05 were considered as statistically significant.
Abbreviations
- NPD1:
-
neuroprotectin D1
- DHA:
-
docosahexaenoic acid
- NF-κB:
-
Nuclear factor kappa B
- cIAP:
-
inhibitor of apoptosis
- BIRC:
-
baculoviral IAP repeat-containing proteins
- OS:
-
oxidative stress
- DISC:
-
death-inducing signaling complex
- PLA2:
-
phospholipase A2
- 15-LOX-1:
-
15-lipoxygenase-1
- COX-2:
-
cyclooxygenase-2
- RPE:
-
retinal pigment epithelial cells
- LXA4:
-
lipoxin A4
- TNF-α:
-
tumor necrosis factor alpha
- Nec1:
-
Necrostatin 1
- RIP1:
-
receptor interacting protein 1
- TAK1:
-
TGFβ-activated kinase 1
- TAB:
-
(TAK1)-binding protein
- TGFβ:
-
transforming growth factor beta
- IKK:
-
I kappa B kinase
- TRAF:
-
tumor necrosis factor receptor-associated factor
- TNFR1:
-
tumor necrosis factor receptor 1
References
Han J, Zhong CQ, Zhang DW . Programmed necrosis: backup to and competitor with apoptosis in the immune system. Nat Immunol 2011; 12: 1143–1149.
Gyrd-Hansen M, Meier P . IAPs: from caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat Rev Cancer 2010; 10: 561–574.
Mace PD, Shirley S, Day CL . Assembling the building blocks: structure and function of inhibitor of apoptosis proteins. Cell Death Differ 2010; 17: 46–53.
Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N et al. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal 2007; 19: 2056–2067.
Huang JF, Shang L, Zhang MQ, Wang H, Chen D, Tong JB et al. Differential neuronal expression of receptor interacting protein 3 in rat retina: involvement in ischemic stress response. BMC Neurosci 2013; 14: 16.
Nicotera P, Lipton SA . Excitotoxins in neuronal apoptosis and necrosis. J Cereb Blood Flow Metab 1999; 19: 583–591.
Rosenbaum DM, Degterev A, David J, Rosenbaum PS, Roth S, Grotta JC et al. Necroptosis, a novel form of caspase-independent cell death, contributes to neuronal damage in a retinal ischemia-reperfusion injury model. J Neurosci Res 2010; 88: 1569–1576.
Long JS, Ryan KM . New frontiers in promoting tumour cell death: targeting apoptosis, necroptosis and autophagy. Oncogene 2012; 31: 5054–5060.
Bazan NG, Molina MF, Gordon WC . Docosahexaenoic acid signalolipidomics in nutrition: significance in aging, neuroinflammation, macular degeneration, Alzheimer's, and other neurodegenerative diseases. Annu Rev Nutr 2011; 31: 321–351.
Calandria JM, Marcheselli VL, Mukherjee PK, Uddin J, Winkler JW, Petasis NA et al. Selective survival rescue in 15-lipoxygenase-1-deficient retinal pigment epithelial cells by the novel docosahexaenoic acid-derived mediator, neuroprotectin D1. J Biol Chem 2009; 284: 17877–17882.
Marcheselli VL, Mukherjee PK, Arita M, Hong S, Antony R, Sheets K et al. Neuroprotectin D1/protectin D1 stereoselective and specific binding with human retinal pigment epithelial cells and neutrophils. Prostaglandins Leukot Essent Fatty Acids 2010; 82: 27–34.
Mukherjee PK, Marcheselli VL, Serhan CN, Bazan NG . Neuroprotectin D1: a docosahexaenoic acid-derived docosatriene protects human retinal pigment epithelial cells from oxidative stress. Proc Natl Acad Sci USA 2004; 101: 8491–8496.
Mukherjee PK, Marcheselli VL, Barreiro S, Hu J, Bok D, Bazan NG . Neurotrophins enhance retinal pigment epithelial cell survival through neuroprotectin D1 signaling. Proc Natl Acad Sci USA 2007; 104: 13152–13157.
Calandria JM, Mukherjee PK, de Rivero Vaccari JC, Zhu M, Petasis NA, Bazan NG . Ataxin-1 poly(Q)-induced proteotoxic stress and apoptosis are attenuated in neural cells by docosahexaenoic acid-derived neuroprotectin D1. J Biol Chem 2012; 287: 23726–23739.
Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM . ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res 1996; 62: 155–169.
Strauss O . The retinal pigment epithelium in visual function. Physiol Rev 2005; 85: 845–881.
Sparrow JR, Hicks D, Hamel CP . The retinal pigment epithelium in health and disease. Curr Mol Med 2010; 10: 802–823.
Lukiw WJ, Cui JG, Marcheselli VL, Bodker M, Botkjaer A, Gotlinger K et al. A role for docosahexaenoic acid-derived neuroprotectin D1 in neural cell survival and Alzheimer disease. J Clin Invest 2005; 115: 2774–2783.
Zhao Y, Calon F, Julien C, Winkler JW, Petasis NA, Lukiw WJ et al. Docosahexaenoic acid-derived neuroprotectin D1 induces neuronal survival via secretase- and PPARγ-mediated mechanisms in Alzheimer's disease models. PLoS ONE 2011; 6: e15816.
Serhan CN, Krishnamoorthy S, Recchiuti A, Chiang N . Novel anti-inflammatory—proresolving mediators and their receptors. Curr Top Med Chem 2011; 11: 629–647.
Serhan CN, Yang R, Martinod K, Kasuga K, Pillai PS, Porter TF et al. Maresins: novel macrophage mediators with potent antiinflammatory and proresolving actions. J Exp Med 2009; 206: 15–23.
Viatour P1, Merville MP, Bours V, Chariot A . Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem Sci 2005; 30: 43–52.
Hong SY, Yoon WH, Park JH, Kang SG, Ahn JH, Lee TH et al. Involvement of two NF-kappa B binding elements in tumor necrosis factor alpha -, CD40-, and epstein-barr virus latent membrane protein 1-mediated induction of the cellular inhibitor of apoptosis protein 2 gene. J Biol Chem 2000; 275: 18022–18028.
Eckelman BP, Salvesen GS . The human anti-apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J Biol Chem 2006; 281: 3254–3260.
Xiao G, Fu J . NF-κB and cancer: a paradigm of Yin-Yang. Am J Cancer Res 2011; 1: 192–221.
Hoffmann A, Levchenko A, Scott ML, Baltimore D . The IκB-NF-κB signaling module: temporal control and selective gene activation. Science 2002; 298: 1241–1245.
Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE et al. Oscillations in NF-κB signaling control the dynamics of gene expression. Science 2004; 306: 704–708.
Lipniacki T, Paszek P, Brasier AR, Luxon B, Kimmel M . Mathematical model of NFkappaB regulatory module. J Theor Biol 2004; 228: 195–215.
Sung MH, Salvatore L, De Lorenzi R, Indrawan A, Pasparakis M, Hager GL et al. Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PLoS ONE 2009; 4: e7163.
Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463.
He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 11001111.
Nicotera P, Melino G . Regulation of the apoptosis-necrosis switch. Oncogene 2004; 23: 2757–2765.
Marcheselli VL, Hong S, Lukiw WJ, Tian XH, Gronert K, Musto A et al. Novel docosanoids inhibit brain ischemia-reperfusion-mediated leukocyte infiltration and pro-inflammatory gene expression. J Biol Chem 2003; 278: 43807–43817.
Belayev L, Khoutorova L, Atkins KD, Eady TN, Hong S, Lu Y et al. Docosahexaenoic acid therapy of experimental ischemic stroke. Transl Stroke Res 2011; 2: 33–41.
Pizzi M, Sarnico I, Lanzillotta A, Battistin L, Spano PF . Post-ischemic brain damage: NF-κB dimer heterogeneity as a molecular determinant of neuron vulnerability. FEBS J 2009; 276: 27–35.
Valerio A, Dossena M, Bertolotti P, Boroni F, Sarnico I, Faraco G et al. Leptin is induced in the ischemic cerebral cortex and exerts neuroprotection through NF-kappaB/c-Rel-dependent transcription. Stroke 2009; 40: 610–617.
Jiang C, Xuan Z, Zhao F, Zhang MQ . TRED: a transcriptional regulatory element database, new entries and other development. Nucleic Acids Res 2007; 35: D137–D140.
Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 1998; 26: 362–367.
Tsunoda T, Takagi T . Estimating transcription factor bindability on DNA. Bioinformatics 1999; 15: 622–630.
Bren GD, Solan NJ, Miyoshi H, Pennington KN, Pobst LJ, Paya CV et al. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene 2001; 20: 7722–7733.
Ueberla K, Lu Y, Chung E, Haseltine WA . The NF-kappa B p65 promoter. J Acquir Immune Defic Syndr 1993; 6: 227–230.
Viswanathan M, Yu M, Mendoza L, Yunis JJ . Cloning and transcription factor-binding sites of the human c-rel proto-oncogene promoter. Gene 1996; 170: 271–276.
Tae G, Scatena M, Stayton PS, Hoffman AS . PEG-cross-linked heparin is an affinity hydrogel for sustained release of vascular endothelial growth factor. J Biomater Sci Polym Ed 2006; 17: 187–197.
Acknowledgements
This work was supported by National Institutes of Health (NIH) to NGB grants R01 EY005121 (National Eye Institute to NGB), the Eye, Ear, Nose & Throat Foundation of New Orleans and P30 GM103340 (National Institute of General Medical Sciences to NGB), and in part by the Louisiana Lions Eye Foundation, New Orleans, LA; Research to Prevent Blindness, New York, NY. We thank Professor Tae H Lee from the Department of Biology, College of Science, Yonsei University, South Korea for generously providing the BIRC3 promoter deletion mutants, and Professor Nicos A Petasis from the Department of Chemistry, University of Southern California, USA for providing NPD1 and other lipids.
Author Contributions
JMC and AA contributed equally to the work presented in this paper; JMC and NGB designed experiments; AA and JMC performed assays and analyzed data; AA performed the promoter computational analysis; VB performed the immunohistochemistry; PKM performed the western blot analysis of BIRC3 in MCAo; EK and BKJ performed the LCMS/MS assay of NPD1; LB performed the MCAo experiments; JMC and NGB wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by D Kaplan
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Calandria, J., Asatryan, A., Balaszczuk, V. et al. NPD1-mediated stereoselective regulation of BIRC3 expression through cREL is decisive for neural cell survival. Cell Death Differ 22, 1363–1377 (2015). https://doi.org/10.1038/cdd.2014.233
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.233
This article is cited by
-
cRel and Wnt5a/Frizzled 5 Receptor-Mediated Inflammatory Regulation Reveal Novel Neuroprotectin D1 Targets for Neuroprotection
Cellular and Molecular Neurobiology (2023)
-
Docosanoids Promote Neurogenesis and Angiogenesis, Blood-Brain Barrier Integrity, Penumbra Protection, and Neurobehavioral Recovery After Experimental Ischemic Stroke
Molecular Neurobiology (2018)
-
Effects of DHA on Hippocampal Autophagy and Lysosome Function After Traumatic Brain Injury
Molecular Neurobiology (2018)
-
Cooperation of Rel family members in regulating Aβ1-40-mediated pro-inflammatory cytokine secretion by retinal pigment epithelial cells
Cell Death & Disease (2017)
-
Neuroprotectin D1 upregulates Iduna expression and provides protection in cellular uncompensated oxidative stress and in experimental ischemic stroke
Cell Death & Differentiation (2017)