Abstract
We have previously demonstrated that infection by coxsackievirus B3 (CVB3), a positive-stranded RNA enterovirus, results in the accumulation of insoluble ubiquitin–protein aggregates, which resembles the common feature of neurodegenerative diseases. The importance of protein aggregation in viral pathogenesis has been recognized; however, the underlying regulatory mechanisms remain ill-defined. Transactive response DNA-binding protein-43 (TDP-43) is an RNA-binding protein that has an essential role in regulating RNA metabolism at multiple levels. Cleavage and cytoplasmic aggregation of TDP-43 serves as a major molecular marker for amyotrophic lateral sclerosis and frontotemporal lobar degeneration and contributes significantly to disease progression. In this study, we reported that TDP-43 is translocated from the nucleus to the cytoplasm during CVB3 infection through the activity of viral protease 2A, followed by the cleavage mediated by viral protease 3C. Cytoplasmic translocation of TDP-43 is accompanied by reduced solubility and increased formation of protein aggregates. The cleavage takes place at amino-acid 327 between glutamine and alanine, resulting in the generation of an N- and C-terminal cleavage fragment of ~35 and ~8 kDa, respectively. The C-terminal product of TDP-43 is unstable and quickly degraded through the proteasome degradation pathway, whereas the N-terminal truncation of TDP-43 acts as a dominant-negative mutant that inhibits the function of native TDP-43 in alternative RNA splicing. Lastly, we demonstrated that knockdown of TDP-43 results in an increase in viral titers, suggesting a protective role for TDP-43 in CVB3 infection. Taken together, our findings suggest a novel model by which cytoplasmic redistribution and cleavage of TDP-43 as a consequence of CVB3 infection disrupts the solubility and transcriptional activity of TDP-43. Our results also reveal a mechanism evolved by enteroviruses to support efficient viral infection.
Similar content being viewed by others
Main
Coxsackievirus B3 (CVB3) is a small, positive-stranded RNA enterovirus.1 The single open reading frame of CVB3 is translated into a viral polypeptide that is subsequently cleaved by two virus-encoded proteases 2A and 3C to generate structural and non-structural proteins.2 In addition to processing viral polyprotein, 2A and 3C target host proteins important for maintenance of protein translation and transcription, antiviral activity, and cellular architecture and signaling, contributing to virus-induced pathogenesis.3, 4, 5 Although enteroviral replication takes place exclusively in the cytoplasm, viral infection has been demonstrated to lead to cytoplasmic translocation of nuclear proteins.6 For example, heterogeneous ribonucleoprotein D (hnRNP D) has been shown to translocate from the nucleus to the cytoplasm during enteroviral infection.5, 7, 8 Moreover, hnRNP D is cleaved by 3C and has an antiviral function against enteroviral infection.5, 7, 8 Cytoplasmic translocation after enteroviral infection has also been demonstrated for several other hnRNPs (A1, C, and K);9 however, the significance remains unclear.
Transactive response DNA-binding protein 43 (TDP-43) is an hnRNP that plays a critical role in RNA processing through interacting with ribonucleoprotein complexes in the nucleus to regulate transcription and pre-RNA maturation of selected RNA.10, 11 Previous studies have shown that mutations of TDP-43 are linked with neurodegenerative diseases, in particular, amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD).12, 13, 14, 15, 16, 17 In addition, aberrant cleavage and cytoplasmic aggregation of TDP-43 are identified as molecular signatures for most forms of ALS and FTLD and contribute significantly to disease progression.18 However, the role of TDP-43 in virus-induced diseases has not been studied. Recent evidence suggests that CVB3-induced pathogenesis resembles the pathological features of neurodegenative disease, that is, abnormal accumulation of insoluble, misfolded protein aggregates (also known as proteinopathies),3, 19, 20 prompting us to hypothesize that dysregulation of TDP-43 during CVB3 infection plays a role in viral pathogenesis, and probably viral infectivity as well.
In this study, we demonstrate that CVB3 infection causes a cytoplasmic redistribution and aggregation of TDP-43 in a 2A-dependent manner, followed by its cleavage mediated by 3C, leading to the loss-of-function of native TDP-43. Our data also reveal that the N-terminal cleavage fragment of TDP-43 acts as a dominant-negative mutant that inhibits the biological activity of native TDP-43 in regulating RNA splicing. We further demonstrate a host-protective function for TDP-43 against CVB3 infection. Collectively, our results are the first to show TDP-43 proteinopathies in a model outside of neurodegenerative disease and suggest a mechanism evolved by enteroviruses to promote viral infection and induce viral pathogenesis.
Results
Coxsackievirus B3 infection induces TDP-43 cleavage
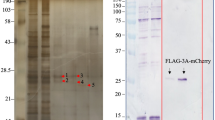
It is well established that TDP-43 proteinopathies contribute to the pathological progression of motor-neuron diseases;12, 13, 14, 15, 16, 17 however, the interaction between virus infection and the host TDP-43 pathway has not yet been explored. In this study, we first examined the protein expression of TDP-43 in various models of CVB3 infection. We demonstrated the production of a previously undetermined immunoreactive fragment of ~35 kDa in A/J mouse heart challenged with 9 days of CVB3 infection (Figure 1a). We then verified this observation in cell culture models. We showed that protein expression of TDP-43 was decreased after CVB3 infection, accompanied by the appearance of a similar 35 kDa band in both primary isolated mouse cardiomyocytes (Figure 1b) and HeLa cells (Figure 1c). To further determine whether the observed fragment is virus-induced, we transiently expressed exogenous HA-TDP-43-GFP in HeLa cells. Following 7 h of CVB3 infection, we showed the generation of a comparable 35 kDa fragment using an anti-HA antibody, suggesting that TDP-43 is targeted for cleavage following CVB3 infection (Figure 1d).
TDP-43 is cleaved following CVB3 infection. (a) TDP-43 expression in A/J mouse heart following 9 days of CVB3 infection. Heart extracts were processed for western blot analysis for protein expression of TDP-43 using an anti-N-terminal TDP-43 antibody. GAPDH level was examined as a protein loading control. (b and c) TDP-43 protein expression in primary mouse cardiomyocytes (b) and HeLa cells (c) infected with CVB3 at an multiplicities of infection (MOI) of 100 and 10, respectively, for various times as indicated. Western blot analysis was conducted as described above for the detection of protein expression of TDP-43, viral capsid protein VP1, and β-actin (loading control). (d) Cleavage of TDP-43 following CVB3 infection. HeLa cells were transiently transfected with a plasmid expressing HA-TDP-43-GFP, followed by 7 h of CVB3 infection. Cells were collected and processed for western blotting using an anti-HA antibody to detect exogenous TDP-43. * indicates a cleaved TDP-43 band at ~35 kDa. cp, cleavage product. Protein levels of pro- and cleaved TDP-43 were quantitated by densitometric analysis using NIH ImageJ, normalized to GAPDH or β-actin, and presented underneath each blot
Viral protease 3C cleaves TDP-43 at amino-acid Q327
To determine whether viral proteases contribute to the cleavage of TDP-43 following CVB3 infection, we carried out an in vitro cleavage assay. Our data showed the production of a similar 35 kDa fragment in HeLa cell lysates upon incubation with viral protease 3C in a time-dependent manner (Figure 2a). We further demonstrated that the catalytically inactive mutant of 3C failed to cleave TDP-43 (Figure 2b). Altogether, our data indicate that 3C is responsible for TDP-43 cleavage.
TDP-43 is cleaved by viral protease 3C. (a and b) Cleavage of TDP-43 by viral protease 3C. (a) HeLa cell lysates (50 μg) were incubated with or without viral protease 3C (0.1 μg) for different times as indicated. (b) Protein extracts from HeLa cells transiently expressing HA-TDP-43-GFP were incubated with wild-type or catalytically inactive mutant of 3C (3Cmut) for 16 h. In vitro cleavage assay was performed as described in the ‘Materials and Methods’. Expression of TDP-43 was examined using an anti-N-terminal TDP-43 (a) or anti-HA antibody (b). (c–e) Effects of general caspase inhibition on CVB3-mediated TDP-43 cleavage. HeLa cells were infected with CVB3 for 7 h (c) or 9 h (e) in the presence or absence of Z-VAD-FMK (zVAD, 50 μM). Western blotting was performed to examine the expression of TDP-43 using an anti-N-terminal TDP-43 antibody and the cleavage of caspase-3 by an anti-caspase-3 antibody (d). VP1 and β-actin was detected as described above. * indicates a cleaved TDP-43 band at ~35 kDa. ‘-‘, vehicle control (treated with DMSO). Densitometric analysis was carried out as in Figure 1
It was previously reported that TDP-43 could be targeted by caspases, resulting in the generation of pathological C-terminal cleavage fragments.21, 22, 23 To eliminate the possibility that the above-detected fragment of TDP-43 is due to caspase-induced cleavage, we pharmacologically inhibited caspase activity using Z-VAD-FMK. Figure 2c showed that treatment with Z-VAD-FMK did not block the generation of the ~35 kDa cleavage products at 7 h post-infection. This is expected as caspase-3 was not activated at this time point (Figure 2d). It was previously shown that CVB3 infection induces caspase activation at a late stage of viral infection.24, 25, 26 We then extended the period of viral infection to 9 h. Figure 2d showed that caspase-3 was activated at this time point as evidenced by the cleavage of caspase-3 and addition of Z-VAD-FMK inhibited this cleavage. As shown in Figure 2e, three cleavage fragments, that is, ~35, ~32, and ~25 kDa, were detected in CVB3-infected cells. The lower two bands were undetected, whereas the upper band accumulated in cells treated with caspase inhibitor (Figure 2e). Collectively, our results suggest that the production of the ~35 kDa bands is not a result of caspase activation.
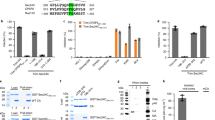
Using netpicoRNA V1.0 algorithm, several potential cleavage sites on TDP-43 were predicted. On the basis of the predicted sites, point mutants of TDP-43 were constructed by site-directed mutagenesis. Among these mutants, TDP-43-Q327L, in which the glutamine (Q) at position 327 was replaced with leucine (L), was cleavage resistant, indicating TDP-43 is cleaved at 327 (Figure 3a). In vitro cleavage assay further confirmed that TDP-43-Q327L failed to be cleaved by 3C, whereas wild-type TDP-43 was proteolytically processed by 3C, but not 2A (Figure 3b). Collectively, our data suggest that TDP-43 is cleaved by 3C at Q327, generating a 35 kDa N-terminal (TDP-43-N) and an 8-kDa C-terminal cleavage fragment (TDP-43-C) (Figure 3c).
TDP-43 is cleaved at Q327. (a) HeLa Cells transiently transfected with HA-TDP-43-GFP (WT) or HA-TDP-43Q327L-GFP (Q327L) were either sham or CVB3-infected for 7 h. Cells were collected for western blot analysis of the expression of TDP-43 using anti-HA antibody, VP1, and β-actin. Densitometric analysis was carried out as in Figure 1. (b) Fifty microgram of protein extracts from HeLa cells transiently expressing WT or HA-TDP-43Q327L-GFP was incubated with increasing concentrations of viral protease 3C or 2A as indicated for 8 h. CVB3-infected HeLa lysates were used as a control to compare the generation of similar 35 kDa cleavage fragments. Protein expression of TDP-43, VP1, and β-actin was detected as described above. * indicates a cleaved TDP-43 band at ~35 kDa. (c) Schematic diagram of the functional domains, the identified cleavage site, and the resulting cleavage fragments of TDP-43. RRM, RNA-recognition motif; NLS, nuclear localization signal; and NES, nuclear export signal
Viral protease 2A is responsible for cytoplasmic translocation and aggregation of TDP-43
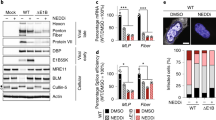
Cytoplasmic translocation and aggregation of TDP-43 are the pathological hallmarks for ALS and FTLD.18 To understand the effect of CVB3 infection on TDP-43 localization, we utilized a fluorescent mCherry tagged TDP-43. As shown in Figure 4a, TDP-43 was localized exclusively in the nucleus in sham-infected cells, upon infection TDP-43 was translocated from the nucleus to the cytosol. In neurodegenerative disease, increasing evidence has shown that cytoplasmic TDP-43 localizes to stress granules (SGs) to form protein aggregates.27, 28, 29 SGs are cytoplasmic aggregates of stalled translation initiation complex.30 Here we used a construct expressing EGFP-tagged G3BP1, a core component of SGs, to monitor the formation of SGs. Similar to our previous findings,5 CVB3 infection induced the formation of SGs at 3 h post-infection (Figure 4a). Interestingly, we found that ~83% of cells infected with CVB3 exhibited punctate cytoplasmic staining patterns of TDP-43, co-localizing to SG aggregates (Figure 4a). We further determined whether viral proteases are responsible for the translocation of TDP-43. Figure 4b demonstrated that transient expression of 2A, but not 3C, elicited significant cytoplasmic redistribution and aggregate formation of TDP-43, similar to those observed during viral infection, indicating that 2A expression is sufficient and responsible for cytoplasmic translocation and aggregation of TDP-43 during CVB3 infection.
Viral protease 2A is responsible for cytoplasmic relocalization and aggregation of TDP-43 during CVB3 infection. (a) Cytoplasmic redistribution and aggregation of TDP-43 after CVB3 infection. HeLa cells transiently co-expressing HA-TDP-43-mCherry and EGFP-G3BP1 were sham- or CVB3-infected for 3 h. Confocal fluorescence images are presented (TDP-43, red; G3BP1, green; nucleus, DAPI (blue)). Percentage of cells expressing cytoplasmic TDP-43 was quantified over 10 different images as described in ‘Materials and Methods’ (b) Localization of TDP-43 after viral protease expression. HeLa cells were transiently co-transfected with HA-TDP-43-GFP and viral protease 2A, 3C, or empty vector for 24 h. Immunostaining was carried out for the detection of TDP-43 using anti-HA antibody (Alexa-fluor-594, red). Cell nuclei were counterstained with DAPI (blue). Percentages of TDP-43-positive cells expressing cytoplasmic TDP-43 in 10 different images are shown to the right
The solubility of TDP-43 is reduced following CVB3 infection
It was previously reported that, under disease condition, the solubility of TDP-43 is changed and altered solubility is correlated with cytoplasmic translocation and aggregation of TDP-43 and disease progression.17, 31, 32 To determine whether CVB3 infection affects TDP-43 solubility, cells were infected with CVB3 for 7 h and cellular proteins were extracted sequentially using RIPA and then urea buffers. We found that, in sham-infected cells, TDP-43 was mainly present in RIPA-soluble fraction, whereas in CVB3-infected cells, both native and cleavage forms of TDP-43 were transferred to the RIPA-insoluble, urea-soluble fraction (Figure 5), suggesting that the solubility of TDP-43 is decreased after CVB3 infection.
TDP-43 solubility is reduced in cells infected by CVB3. HeLa cells were infected with CVB3 for 7 h, cell lysates were collected and proteins were sequentially extracted using RIPA and Urea buffers. Western blotting was performed using an anti-N-terminal TDP-43 antibody for the detection of full-length and the N-terminal cleaved form of TDP-43. Expression of GAPDH was examined to evaluate the purity of protein extraction (GAPDH is presented in RIPA-soluble fraction) and as a loading control. * indicates the N-terminal fragment of TDP-43. RIPA-S, RIPA buffer-soluble; RIPA-I, RIPA buffer-insoluble. Densitometric analysis was carried out as in Figure 1
TDP-43-N localizes to stress granules to form protein aggregates whereas TDP-43-C is rapidly degraded via the proteasomal pathway
To explore the possible significance or consequence of TDP-43 cleavage during CVB3 infection, we cloned the human TDP-43-N- (amino acids 1–327) and C- (amino acids 328–414) terminal cleavage products into a vector expressing an HA tag. Western blotting verified the protein expression of full-length and N-terminal TDP-43 after transient transfection (Figure 6a). However, TDP-43-C failed to be detected following transfection (Figure 6a). Previous studies have demonstrated degradation of caspase-induced cleavage products of TDP-43 by the proteasome.33 To determine whether TDP-43-C is rapidly degraded following transient transfection, HeLa cells were treated with bafilomycin or MG132 to inhibit autophagic and proteasomal pathways, respectively. Figure 6b showed that treatment with MG132, but not bafilomycin, rescued TDP-43-C protein expression. This data suggest that the C-terminal fragment is quickly turned over through proteasome-mediated proteolysis, contributing to the loss-of-function of TDP-43 following CVB3 infection. Furthermore, we examined the localization of the cleavage products. TDP-43-C signal was undetectable by confocal microscopy, which is likely owing to its instability (data not shown). TDP-43-N was localized to the nucleus in sham-infected or vector control cells, but translocated to SG aggregates in the cytoplasm upon CVB3 infection or 2A expression (Figure 6c). This result indicates that, in addition to full-length TDP-43 (Figure 4b), the TDP-43-N fragment is also able to form aggregates.
TDP-43-C is rapidly degraded by the proteasome pathway and TDP-43-N localizes to stress granules to form protein aggregates after CVB3 infection. (a) Protein expression of full-length and truncated forms of TDP-43. HeLa cells were transiently transfected with HA-TDP-43-GFP, HA-TDP-43-N, or HA-TDP-43-C as indicated. Protein expression of various types of TDP3 was detected using an anti-HA antibody. The level of β-actin was examined as a loading control. (b) TDP-43-C is degraded through the proteasome pathway. HeLa cells were transiently transfected with HA-TDP-43-C in the presence of 200 nM bafilomycin (a lysosome inhibitor), 1 μM MG132 (a proteasome inhibitor), or equal volume of DMSO as a vehicle control for 24 h. Western blotting was performed using an anti-C-terminal TDP-43 antibody which detects both endogenous TDP-43 (~43 kDa) and exogenous HA-TDP-43-C (~8 kDa). Densitometric analysis of TDP-43 proform, TDP-43-N and TDP-43-C was carried out as in Figure 1. (c) Cytoplasmic localization and aggregation of TDP-43-N following either CVB3 infection or viral protease 2A expression. HeLa cells were transiently co-transfected with HA-TDP-43-N and EGFP-G3BP1 for 24 h, followed by either CVB3 infection for 3 h or second-round of transfection with viral protease 2A as indicated. Immunostaining was performed for the detection of TDP-43-N using anti-HA antibody (Alexa-fluor-594, red). Cell nuclei were counterstained with DAPI (blue). GFP signal of EGFP-G3BP1 is shown in green. Percentage of cells expressing cytoplasmic TDP-43 was quantified as in Figure 4
TDP-43-N compromises the function of native TDP-43 in CFTR exon 9 skipping
TDP-43 plays an important role in the regulation of alternative splicing.34, 35, 36 To determine whether TDP-43-N has an impact on the function of native TDP-43 in RNA splicing, we utilized a well-established technique, CFTR exon 9 skipping.17, 34 TDP-43 facilitates exon 9 skipping by interacting with UG repeats in intron 8 of CFTR pre-mRNA, thus generating an exon 9-deficient transcript at resting states.10, 11, 34 HeLa cells were transiently transfected with a CFTR minigene reporter construct (TG(13)T(5)),17, 34 together with empty vector, full-length TDP-43, or TDP-43-N. Transfection was carried out for 24 h to achieve desired expression levels of TDP-43-N similar to those observed in virus-infected cells. RT-PCR was performed to detect the transcripts of CFTR reporter plasmid. We found that expression of TDP-43-N resulted in a significant decrease in CFTR exon 9 skipping (decreased ratio of spliced to unspliced exon 9) relative to vector controls (Figure 7). These results suggest that the N-terminal cleavage fragment of TDP-43 functions as a dominant-negative mutant that inhibits the activity of native TDP-43 in regulating RNA splicing. It was noted that transient transfection of full-length TDP-43 did not further increase CFTR splicing compared with empty-vector control (Figure 7), in contrast to an early report.22 The discrepancy is likely due to the differences in the basal levels of endogenous TDP43 activity and the amount of exogenous TDP43 transfected between two studies. The endogenous TDP-43 exon skipping activity in our study appeared to be already high (higher than that in their study22). Thus, modest increase of exogenous TDP43 due to a shorter term of transfection (24 h in our study versus 48 h in their report22) may not be sufficient to promote further increase in exon 9 exclusion. Our result is consistent with an early report with the same period of TDP-43 transfection (24 h) showing that exon skipping activity in cells expressing exogenous TDP43 is comparable to cells expressing empty-vector, but significantly higher compared with cells depleted of TDP-43.17
TDP-43-N compromises the function of native TDP-43 in CFTR splicing. HeLa cells were transiently co-transfected with CFTR reporter construct (TG(13)T(5)) with empty vector control, full-length TDP-43, or TDP-43-N for 24 h. RNA was extracted and RT-PCR was performed to examine CFTR exon 9 skipping. PCR products were visualized on 1.5% agarose gels (upper panel) and quantified by NIH imageJ software and presented as ratios of spliced to unspliced exon 9 (mean±S.D., n=3; lower panel). **P<0.01. Arrows indicate previously documented splicing variants. Exon 9 inclusion (+) and exclusion (−) are shown
TDP-43 knockdown promotes CVB3 replication
To determine whether TDP-43 plays a role in CVB3 infection, HeLa cells were treated with siRNA specific for TDP-43 mRNA, followed by viral infection. Figures 8a and b showed that gene-silencing of TDP-43 resulted in an approximately 2-fold increase in virus titers as compared with control, indicating a host-protective effect of TDP-43 against CVB3 infection. To further understand the potential mechanism by which knockdown of TDP-43 enhances viral replication, immunostaining was performed to determine whether TDP-43 interacts directly with the viral RNA. We utilized a double-stranded RNA (dsRNA) antibody to monitor the accumulation of viral replication intermediates. Confocal microscope images and Pearson’s correction analysis showed that TDP-43 signal did not significantly co-localize with dsRNA staining (Figure 8c), suggesting that the antiviral activity of TDP-43 may not be through direct binding with CVB3 RNA and subsequent interfering with its replication, rather than an indirect mechanism.
Knockdown of TDP-43 enhances CVB3 replication. HeLa cells were transfected with an siRNA targeting TDP-43 (siTDP-43) or a scramble siRNA (siCon) for 48 h followed by CVB3 infection for 16 h at an multiplicities of infection (MOI) of 1. (a) Western blotting was conducted to examine protein expression of TDP-43 and β-actin (loading control). Densitometric analysis was performed as in Figure 1. * indicates the N-terminal cleavage fragment of TDP-43. (b) Plaque assay was carried out to assess viral titers in the supernatant. Viral titers were expressed as plaque formation units (pfu) per ml and presented as mean±S.D. (n=3), *P<0.05. (c) Localization of TDP-43 and dsRNA in the cytoplasm of CVB3-infected cells. HeLa cells were transfected with HA-TDP-43 for 24 h, followed by CVB3 infection for 3 h. Immunostaining was carried out for the detection of TDP-43 using anti-HA antibody (green) and dsRNA using anti-dsRNA antibody (red). Cell nuclei were counterstained with DAPI (blue). Pearson's Correlation Coefficient (PCC) was calculated using Volocity software Version 5.2.1 over at least three images
Discussion
In this study, we provide the first evidence that TDP-43 is manipulated by enteroviruses to contribute to viral pathogenesis and benefit viral replication. TDP-43 is a nuclear-cytoplasmic shuttling protein passing across the nuclear membrane through the activity of its intrinsic nuclear localization signal (NLS) and nuclear export signal. However, under normal conditions, TDP-43 is predominantly localized to the nucleus. In neurodegenerative diseases, patients suffering from ALS or FTLD exhibit cytoplasmic translocation of TDP-43.14, 15, 17, 18, 31 In this study, we showed a similar translocation pattern of TDP-43 from the nucleus to the cytoplasm following CVB3 infection. As enteroviral protease 2A was previously shown to cleave key components of the nuclear pore complex, resulting in the disruption of nuclear import pathways and consequent cytoplasmic redistribution of selective nuclear proteins, including several members of the hnRNPs,9, 37, 38, 39 we asked whether expression of 2A is sufficient to induce similar cytoplasmic translocation patterns of TDP-43. Our data indicate that transient expression of 2A in cells recapitulates this phenomenon, indicating that 2A is responsible for the cytoplasmic translocation of TDP-43. Identification of TDP-43 as another member of hnRNPs undergoing cytoplasmic relocalization during CVB3 infection suggests that modulation of hnRNP distribution may be a common strategy exploited by enteroviruses to allow for efficient viral production. Although 3C can directly cleave TDP-43 and the cleavage products tends to promote aggregation, expression of 3C alone was unable to induce cytoplasmic translocation and aggregation of TDP-43. This is because the absence of viral infection or viral protease 2A, TDP-43 remains in the nucleus, inaccessible to 3C that is mainly present in the cytoplasm.
TDP-43 proteinopathies are characterized by reduced solubility and increased cytoplasmic accumulation of TDP-43 as ubiquitinated protein aggregates.18 Here we demonstrated that CVB3 infection causes a decrease in solubility of both native and the N-terminal cleaved form of TDP-43. These findings are accompanied by the observations that CVB3 infection or ectopic expression of 2A induces cytoplasmic redistribution and aggregation of full-length and N-terminal TDP-43. In addition to reduced solubility, a number of studies have shown that mutated TDP-43 and caspase-derived C-terminal TDP-43 fragments that are often detected in the brain of ALS and FTLD patients are highly prone to aggregation.17, 22, 31, 32 Furthermore, it was reported that wild-type TDP-43 and cytoplasmically restricted TDP-43 (mutated at its NLS) are also able to form protein aggregates in the cytoplasm, which are associated with increased protein phosphorylation and ubiquitination, and co-localized with SG markers.29, 40 The exact mechanism leading to decreased solubility and increased aggregation of TDP-43 following CVB3 infection is still not known, but could be related to cytoplasmic translocation of TDP-43, which allows for direct interaction of the two RNA-recognition motifs (RRM 1 and 2) on full-length TDP-43 and TDP-43-N, with cytosolic SGs containing various RNA transcripts and proteins that are involved in protein ubiquitination and phosphorylation.
SGs are dynamic cytoplasmic aggregates of stalled initiation complexes that play an antiviral role against enteroviral infection.5, 41, 42 We have previously demonstrated that shutoff of host protein translation upon CVB3 infection triggers the formation of SGs at ~3 h post infection and protease 3C-mediated cleavage of G3BP1 contributes, at least in part, to SG disassembly at ~5 h post-infection.5 Similar to these findings, cytosolic aggregates of TDP-43 are formed and co-localize to SGs at ~3 h (Figure 4a) and then disassembled at late stage of viral infection (data not shown). Cytoplasmic TDP-43 has been identified as a component of SGs in response to stress; it is thus plausible that formation and disappearance of TDP-43 aggregates are the consequences of SG assembly and disassembly. Whether TDP-43 also plays a role in coxsackieviral modulation of the formation and disassembly of SGs and the significance of TDP-43 accumulation in SGs are still unknown and warrant future investigations.
Although both the ubiquitin–proteasome system and autophagy are implicated to play a role in the disposal of TDP-43, TDP-43 is a relatively stable protein with a half-life between 12 and 34 h depending on the cell types.18 In this study, we found that the 3C-induced TDP-43-C is highly labile and undetectable in cells either infected by CVB3 or transiently expressing TDP-43-C. We further demonstrated that TDP-43-C is rapidly degraded through the proteasome pathway as treatment with the proteasome inhibitor restores the protein level of TDP-43-C. This finding is consistent with previous observation that caspase-induced C-terminal truncation product of TDP-43 has a much shorter half-life (~4 h) compared with full-length TDP-43.18 The molecular basis leading to increased turnover of TDP-43-C remains unclear. It is postulated that cleavage may cause conformational change of the TDP-43-C, which enables it for direct and quick access to the chamber of the 20S proteasome.43 As TDP-43-C contains a portion of the glycine-rich region that is required for its interaction with other hnRNPs, and for the regulation of alternative splicing and transcriptional activity,12 we propose that rapid removal of TDP-43-C contributes to the loss-of-function of TDP-43 following CVB3 infection.
In addition to loss-of-function, our results also suggest a gain-of-toxic-function for TDP-43-N. We demonstrated that expression of TDP-43-N in HeLa cells, which contain abundant endogenous TDP-43, results in a significant reduction of CFTR splicing, indicating that TDP-43-N inhibits the biological functions of native TDP-43 as a dominant-negative mutant. TDP-43-N comprises both RNA-recognition motifs (RRM) 1 and 2, but lacks the C-terminal glycine-rich region that plays a key role in modulating RNA splicing.12 Thus, we speculate that TDP-43-N negatively regulates the activity of native TDP-43 by competing for substrate RNA binding without being able to regulate RNA maturation. However, the significance of TDP-43 function during viral infection remains unknown.
Our finding that knockdown of TDP-43 results in a significant increase in viral titers suggests an antiviral function for TDP-43 against CVB3 infection. Like other viral pathogens, enteroviruses have evolved to manipulate host proteins to provide a favorable environment for productive viral infections. Among them, several members of the hnRNPs family have been previously shown to play a role in regulating viral replication.7, 8, 39, 44, 45, 46, 47 For examples, hnRNP C and K were documented to interact with the non-coding regions of enteroviral RNA to facilitate translation initiation and RNA synthesis.44, 45, 47 Similar to our findings in the current study, enteroviral infection causes cytoplasmic redistribution and cleavage of hnRNP D and deletion of this protein results in enhanced viral replication, suggesting that hnRNA D acts as a host restriction factor against enterovirus infection.7, 8, 39 The antiviral mechanisms of hnRNP D appear to involve negative regulation of internal ribosome entry site (IRES)-mediated translation of viral RNA via direct interaction with the IRES of enteroviruses,7, 46 and viral RNA degradation by targeting the 3′-untranslated region (UTR) of enteroviral genome.39 The exact mechanism of TDP-43 suppressing CVB3 replication remains to be clarified. In this study, we tested our hypothesis that TDP-43 interacts directly with the viral RNA through its RRM domains, leading to the inhibition of viral translation and RNA replication. However, our data do not support this notion as TDP-43 and viral RNA intermediates do not appear to co-localize, indicating an indirect mechanism of TDP-43 action. We speculate that TDP-43 inhibits CVB3 replication by regulating its host RNA substrates that have either pro- or antiviral functions, instead of direct targeting on viral genome. It is important to note that TDP-43 functions as a complex of transcriptional regulators and hnRNPs. Thus, we argue that single knockdown of TDP-43 may only moderately ameliorate viral replication as observed in the current study, whereas collective modulation of multiple hnRNPs would yield a greater impact on viral replication.
On the basis of the results in the present study, we propose a model by which CVB3 manipulates the TDP-43 pathway, contributing to viral pathogenesis (Figures 9a–e). Under normal conditions, TDP-43 participates in regulating RNA maturation in the nucleus. Upon CVB3 infection, viral protease 2A induces redistribution of TDP-43 from the nucleus to the cytoplasm (Figure 9a), where TDP-43 is targeted by viral protease 3C for cleavage at Q327 to yield two previously undiscovered cleavage fragments, TDP-43-N and TDP-43-C (Figure 9b). Cytoplasmic translocation of TDP-43 causes decreased solubility and increased aggregation of TDP-43 (Figure 9c). The TDP-43-C fragment is rapidly degraded through the proteasomal pathway, leading to its loss-of-function (Figure 9d), whereas TDP-43-N is prone to form aggregates (Figure 9c) and negatively regulates the function of native TDP-43 in alternative RNA splicing, possibly through competing for substrate RNA binding (Figure 9e). Taken together, our findings are the first to document enteroviral manipulation of TDP-43 during infection, leading to a phenotype similar to TDP-43 proteinopathies observed in neurodegenerative diseases.
Proposed model of CVB3 manipulation of TDP-43 pathway. Nuclear TDP-43 participates in the regulation of RNA metabolism under normal condition. Upon CVB3 infection, viral protease 2A causes the translocation of TDP-43 from the nucleus to the cytoplasm (a), where TDP-43 is cleaved at amino-acid Q327 by viral protease 3C to generate TDP-43-N and TDP-43-C (b). The solubility of cytoplasmic full-length TDP-43 and TDP-43-N is reduced and prone to aggregation (c). TDP-43-C is rapidly degraded via the proteasomal degradation pathway, leading to its loss-of-function (d), whereas TDP-43-N functions as a dominant-negative mutant, which inhibits the biological activity of TDP-43 in RNA splicing (e)
Materials and Methods
Cell culture
HeLa cells (American Type Culture Collection) were grown in Dulbecco’s modified Eagle’s medium (DMEM, Thermo, #SH30243.01) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Life Technologies, Grand Island, NY, USA; #12483-020). Isolation and culture of primary neonatal mouse cardiomyocytes were performed as described previously.48 Briefly, neonatal cardiomyocytes were obtained by enzymatic dissociation of cardiac ventricles from 1–2-day-old A/J mouse neonates. Non-myocytes were removed via two rounds of pre-plating on culture dishes and cytosine 1-β-D-arabinofuranoside (Sigma-Aldrich, St. Louis, MO, USA; #C1768) was added to inhibit the growth of contaminating non-myocytes. The enriched cardiomyocytes were cultured in DMEM with 10% FBS and 10% horse serum.
Viral infection and drug treatment
CVB3 infections of cardiomyocytes and HeLa cells were performed by incubating cells with CVB3 (Kandolf strain) at multiplicities of infection of 10, unless otherwise specified, in serum-free DMEM. After 1 h (h) infection, the medium containing virus was replaced with fresh complete DMEM. Sham-infected cells were treated with equal volumes of phosphate-buffered saline (PBS).
CVB3 infection of A/J mice was performed as previously described.49 Briefly, male A/J mice (Jackson Laboratory, Sacramento, CA, USA; #000646) at ~5 weeks of age were either infected intraperitoneally with 105 plaque-forming units (pfu) of CVB3 or sham infected with PBS. After 9 days of viral infection, mouse hearts were harvested for western blot analysis of protein expression of TDP-43. These studies were performed in strict accordance with the recommendation in the Guide for the Care and Use of Laboratory Animals of the Canadian Council on Animal Care and were approved by the Animal Care Committee at the University of British Columbia.
For caspase inhibition experiments, Z-VAD-FMK (BD Biosciences, San Jose, CA, USA; #550377), a general caspase inhibitor, was added to the medium after 1 h viral infection. For the study of the degradation of TDP-43 cleavage fragments, cells were treated with 1 μM proteasome inhibitor MG132 (Sigma-Aldrich, #C2211) or 200 nM lysosome inhibitor bafilomycin A1 (Sigma-Aldrich, #B1793) for 16 h prior to collection. Control cells were treated with equal volumes of dimethyl sulfoxide (DMSO).
Plasmid construction, siRNAs and transient transfection
The HA-tagged TDP-43 construct was used as the template to generate the HA-TDP-43-N1-327 and HA-TDP-43-C328-414 constructs. The HA-TDP-43Q327L-GFP mutant was established to replace the glutamine (Q) of wild-type TDP-43 at amino-acid 327 with leucine (L). The pIRES-2A and pIRES-3C constructs were generated as previously described.5 Briefly, the open reading frame of CVB3-2A was cloned into the pIRES vector (Clontech, #631605) using BamHI and SalI restriction cut sites, whereas CVB3-3C was inserted into the XbaI and SalI restriction sites. The EGFP-tagged Ras-GTPase-activating protein (GAP) SH3 domain binding protein-1 (G3BP1) construct was a generous gift from Dr. Jamal Tazi at the Institut de Génétique Moléculaire de Montpellier (Montpellier cedex 5, France). The small interfering RNA (siRNA) against human TDP-43 was obtained from Dharmacon (#L-012394). For transfection, cells were transiently transfected with cDNA constructs and siRNAs using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA; #11668019) and Oligofectamine (Invitrogen, #12252-011), respectively, for 24 or 48 h following the manufacturer’s protocols.
Western blot analysis
After CVB3 infection or transfection, cells were harvested using modified oncogen science lysis buffer (250 mM NaCl, pH 7.2, 50 mM Tris-HCl, 0.1% NP-40, 2 mM EDTA, and 10% glycerol, supplemented with protease inhibitors). Protein quantification was performed by Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA; #500-0006). Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes. The blots were processed using standard techniques as previously described,5 and immunoreactive bands were visualized by enhanced chemiluminescence. The primary antibodies used in this study are: anti-N-terminal TDP-43 (Cedarlane, Surrey, BC, Canada; #10782-2-AP), anti-C-terminal TDP-43 (Sigma-Aldrich, #T1580), anti-HA (Cedarlane, #11867423001), anti-VP1 (Dako, Glostrup, Denmark; #M7064), anti-caspase-3 (Santa Cruz, Dallas, TX, USA; #sc-7272), anti-double-stranded RNA (dsRNA, English and Scientific Consulting Bt), anti-GAPDH (Abcam, Cambridge, UK; #ab9484), and anti-β-actin antibodies (Sigma-Aldrich, #2228). Densitometric analysis was performed using NIH ImageJ software (Open software at http://imagej.nih.gov/ij/index.html), normalized to GAPDH or β-actin, and presented as fold changes compared with sham infection or the first lane as indicated, which was arbitrarily set a value of 1.00.
Cell fractionation
Sequential protein extractions were performed as previously described in order to examine the solubility of TDP-43.17 Briefly, cells were washed twice with PBS, lysed with cold RIPA buffer (Santa Cruz, #sc-24948) supplemented with protease and phosphatase inhibitors. Cell lysates were sonicated and cleared by centrifugation at 100 000 × g for 30 min at 4 °C. Supernatants were collected as RIPA-soluble fractions. RIPA-insoluble protein pellets were washed by resonication and recentrifugation using the same conditions to prevent carry-over. Only supernatants from first extraction were used. RIPA-insoluble pellets were extracted with urea buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris, pH 8.5), followed by sonication and centrifugation at 70 000 × g for 30 min at 22 °C. Supernatants were collected as RIPA-insoluble fractions.
Plaque assay
Virus titer in the cell supernatant was measured using an agar overlay plaque assay as previously described.50 In brief, the samples were serially diluted (10-fold) and overlaid onto a monolayer of HeLa cells. Following 1-h incubation, the medium was replaced with complete DMEM containing 0.75% agar. Three days later, the cells were fixed with Carnoy’s fixature (75% ethanol and 25% acetic acid) and stained with 1% crystal violet. Plaques were counted and virus titers were calculated and expressed as pfu/ml.
CFTR splicing analysis
TDP-43 functional activity was evaluated through measuring exon 9 splicing of cystic fibrosis transmembrane conductance regulator (CFTR) gene as previously documented.17, 34 Briefly, HeLa cells were transiently co-transfected with a CFTR minigene reporter construct (a generous gift from Dr. E Buratti from the International Centre for Genetic Engineering and Biotechnology, Trieste, Italy) and various TDP-43 constructs for 24 h. Total RNA was extracted by QIAGEN RNeasy kit (#74104) and equal amounts of RNA (500 ng) were used for RT-PCR for the detection of exon 9 splicing using primers a2-3 and Bra2 as previously described.17, 34 PCR products were visualized on a 1.5% agarose gel and quantified using ImageJ software.
Indirect immunocytochemical microscopy
Indirect immunofluorescence staining was performed as previously described.51 In brief, after fixation and permeabilization, cells were incubated with primary antibody (anti-HA at a dilution of 1 : 200 or anti-dsRNA at a dilution of 1 : 400) at 4 °C overnight, and then with secondary antibody for 1 h. After washing with PBS, coverslips were mounted using Vectashield mounting medium containing 4’ 6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, #H1200). Images were taken with a Leica SP2 AOBS confocal fluorescence microscope. Quantification of images for cytoplasmic translocation was performed by counting at least 10 images where each image contained five or more cells. Values were presented as % of cells expressing cytoplasmic TDP-43. TDP-43 and dsRNA localization was evaluated using Pearson’s correlation algorithm as described.52, 53
Viral protease purification and in vitro cleavage assay
CVB3 2A, 3C, and catalytically inactive 3C were purified by ion exchange chromatography and size exclusion chromatography as previously described.54, 55 HeLa cell lysates (50 μg) were incubated with various concentrations of purified CVB3 2A, 3C, or 3C mutant as indicated in a cleavage reaction buffer (20 mM HEPES (pH 7.4), 150 mM KOAc and 1 mM DTT) at 37°C for different times as previously described.20, 51 The reaction was stopped by addition of 6 × SDS-PAGE sample buffer and TDP-43 cleavage was assessed by western blot analysis.
Statistical analysis
Quantified results are expressed as means±S.D. Statistical analysis was performed with unpaired Student’s t-test. A value of P<0.05 was considered to be statistically significant. All experiments were repeated at least three times.
Abbreviations
- CVB3, coxsackievirus type B3; TDP-43:
-
transactive response DNA-binding protein-43
- hnRNP:
-
heterogeneous ribonucleoprotein
- FTLD:
-
frontotemporal lobar degeneration
- ALS:
-
amyotrophic lateral sclerosis
- Z-VAD-FMK:
-
benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone
- SG:
-
stress granule
- MG132:
-
N-(benzyloxycarbonyl)leucinylleucinylleucinal
- NLS:
-
nuclear localization signal
- RRM:
-
RNA-recognition motif
- IRES:
-
internal ribosomal entry site
- GAPDH:
-
glyceraldehyde 3-phosphate dehydrogenase
- CFTR:
-
cystic fibrosis transmembrane conductance regulator
References
Esfandiarei M, McManus BM . Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol 2008; 3: 127–155.
Jacobson MF, Baltimore D . Polypeptide cleavages in the formation of poliovirus proteins. Proc Natl Acad Sci USA 1968; 61: 77–84.
Luo H, Wong J, Wong B . Protein degradation systems in viral myocarditis leading to dilated cardiomyopathy. Cardiovasc Res 2010; 85: 347–356.
Huber S, Ramsingh AI . Coxsackievirus-induced pancreatitis. Viral Immunol 2004; 17: 358–369.
Fung G, Ng CS, Zhang J, Shi J, Wong J, Piesik P et al. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS One 2013; 8: e79546.
Gustin KE . Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Res 2003; 95: 35–44.
Cathcart AL, Rozovics JM, Semler BL . Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J Virol 2013; 87: 10423–10434.
Rozovics JM, Chase AJ, Cathcart AL, Chou W, Gershon PD, Palusa S et al. Picornavirus modification of a host mRNA decay protein. MBio 2012; 3: e00431–00412.
Gustin KE, Sarnow P . Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J 2001; 20: 240–249.
Buratti E, Baralle FE . Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 2001; 276: 36337–36343.
Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE . TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem 2005; 280: 37572–37584.
Pesiridis GS, Lee VM, Trojanowski JQ . Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet 2009; 18: R156–R162.
Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol 2008; 63: 535–538.
Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 2008; 40: 572–574.
Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008; 319: 1668–1672.
Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol 2008; 7: 409–416.
Igaz LM, Kwong LK, Chen-Plotkin A, Winton MJ, Unger TL, Xu Y et al. Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. J Biol Chem 2009; 284: 8516–8524.
Lee EB, Lee VM, Trojanowski JQ . Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci 2012; 13: 38–50.
Gao G, Zhang J, Si X, Wong J, Cheung C, McManus B et al. Proteasome inhibition attenuates coxsackievirus-induced myocardial damage in mice. Am J Physiol Heart Circ Physiol 2008; 295: H401–H408.
Shi J, Fung G, Piesik P, Zhang J, Luo H . Dominant-negative function of the C-terminal fragments of NBR1 and SQSTM1 generated during enteroviral infection. Cell Death Differ 2014; 21: 1432–1441.
Nishimoto Y, Ito D, Yagi T, Nihei Y, Tsunoda Y, Suzuki N . Characterization of alternative isoforms and inclusion body of the TAR DNA-binding protein-43. J Biol Chem 2010; 285: 608–619.
Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA 2009; 106: 7607–7612.
Zhang YJ, Xu YF, Dickey CA, Buratti E, Baralle F, Bailey R et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci 2007; 27: 10530–10534.
Carthy CM, Yanagawa B, Luo H, Granville DJ, Yang D, Cheung P et al. Bcl-2 and Bcl-xL overexpression inhibits cytochrome c release, activation of multiple caspases, and virus release following coxsackievirus B3 infection. Virology 2003; 313: 147–157.
Cunningham KA, Chapman NM, Carson SD . Caspase-3 activation and ERK phosphorylation during CVB3 infection of cells: influence of the coxsackievirus and adenovirus receptor and engineered variants. Virus Res 2003; 92: 179–186.
Yuan JP, Zhao W, Wang HT, Wu KY, Li T, Guo XK et al. Coxsackievirus B3-induced apoptosis and caspase-3. Cell Res 2003; 13: 203–209.
Li YR, King OD, Shorter J, Gitler AD . Stress granules as crucibles of ALS pathogenesis. J Cell Biol 2013; 201: 361–372.
Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 2010; 5: e13250.
Parker SJ, Meyerowitz J, James JL, Liddell JR, Crouch PJ, Kanninen KM et al. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem Int 2012; 60: 415–424.
Anderson P, Kedersha N . Stress granules: the Tao of RNA triage. Trends Biochem Sci 2008; 33: 141–150.
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–133.
Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 2006; 351: 602–611.
Zhang YJ, Gendron TF, Xu YF, Ko LW, Yen SH, Petrucelli L . Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol Neurodegener 2010; 5: 33.
Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE . Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 2001; 20: 1774–1784.
Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE . Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res 2005; 33: 6000–6010.
Bose JK, Wang IF, Hung L, Tarn WY, Shen CK . TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem 2008; 283: 28852–28859.
Belov GA, Lidsky PV, Mikitas OV, Egger D, Lukyanov KA, Bienz K et al. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J Virol 2004; 78: 10166–10177.
Park N, Katikaneni P, Skern T, Gustin KE . Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J Virol 2008; 82: 1647–1655.
Wong J, Si X, Angeles A, Zhang J, Shi J, Fung G et al. Cytoplasmic redistribution and cleavage of AUF1 during coxsackievirus infection enhance the stability of its viral genome. FASEB J 2013; 27: 2777–2787.
Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM . Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem 2008; 283: 13302–13309.
Ng CS, Jogi M, Yoo JS, Onomoto K, Koike S, Iwasaki T et al. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol 2013; 87: 9511–9522.
White JP, Cardenas AM, Marissen WE, Lloyd RE . Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2007; 2: 295–305.
Bhattacharyya S, Yu H, Mim C, Matouschek A . Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol 2014; 15: 122–133.
Brunner JE, Nguyen JH, Roehl HH, Ho TV, Swiderek KM, Semler BL . Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J Virol 2005; 79: 3254–3266.
Ertel KJ, Brunner JE, Semler BL . Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J Virol 2010; 84: 4229–4242.
Lin JY, Li ML, Brewer G . mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication. PLoS One 2014; 9: e103827.
Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR . Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5' untranslated region and participates in virus replication. J Gen Virol 2008; 89: 2540–2549.
Ding B, Abe J, Wei H, Huang Q, Walsh RA, Molina CA et al. Functional role of phosphodiesterase 3 in cardiomyocyte apoptosis: implication in heart failure. Circulation 2005; 111: 2469–2476.
Wong J, Zhang J, Yanagawa B, Luo Z, Yang X, Chang J et al. Cleavage of serum response factor mediated by enteroviral protease 2A contributes to impaired cardiac function. Cell Res 2012; 22: 360–371.
Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM et al. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol 2008; 82: 9143–9153.
Shi J, Wong J, Piesik P, Fung G, Zhang J, Jagdeo J et al. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy 2013; 9: 1591–1603.
Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA . Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci 1992; 103: 857–862.
Manders RJ, Koopman R, Sluijsmans WE, van den Berg R, Verbeek K, Saris WH et al. Co-ingestion of a protein hydrolysate with or without additional leucine effectively reduces postprandial blood glucose excursions in Type 2 diabetic men. J Nutr 2006; 136: 1294–1299.
Joachims M, Van Breugel PC, Lloyd RE . Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 1999; 73: 718–727.
Liebig HD, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H et al. Purification of two picornaviral 2A proteinases: interaction with eIF-4gamma and influence on in vitro translation. Biochemistry 1993; 32: 7581–7588.
Acknowledgements
We thank Dr. Eric Jan (University of British Columbia, Canada) for his generous gifts of the purified proteins of coxsackieviral proteases. This work was supported by the Canadian Institutes of Health Research (to HL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by JM Hardwick
Rights and permissions
About this article
Cite this article
Fung, G., Shi, J., Deng, H. et al. Cytoplasmic translocation, aggregation, and cleavage of TDP-43 by enteroviral proteases modulate viral pathogenesis. Cell Death Differ 22, 2087–2097 (2015). https://doi.org/10.1038/cdd.2015.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2015.58
This article is cited by
-
Implications of TDP-43 in non-neuronal systems
Cell Communication and Signaling (2023)
-
Endogenous retroviruses and TDP-43 proteinopathy form a sustaining feedback driving intercellular spread of Drosophila neurodegeneration
Nature Communications (2023)
-
Sublethal enteroviral infection exacerbates disease progression in an ALS mouse model
Journal of Neuroinflammation (2022)
-
TAR DNA-Binding Protein 43 is Cleaved by the Protease 3C of Enterovirus A71
Virologica Sinica (2021)
-
The role of TDP-43 mislocalization in amyotrophic lateral sclerosis
Molecular Neurodegeneration (2020)