Abstract
Emerging evidences suggest that necrosis is programmed and is one of the main forms of cell death in the pathological process in cardiac diseases. Long noncoding RNAs (lncRNAs) are emerging as new players in gene regulation. However, it is not yet clear whether lncRNAs can regulate necrosis in cardiomyocytes. Here, we report that a long noncoding RNA, named necrosis-related factor (NRF), regulates cardiomyocytes necrosis by targeting miR-873 and RIPK1 (receptor-interacting serine/threonine-protein kinase 1)/RIPK3 (receptor-interacting serine/threonine-protein kinase 3). Our results show that RIPK1 and RIPK3 participate in H2O2-induced cardiomyocytes necrosis. miR-873 suppresses the translation of RIPK1/RIPK3 and inhibits RIPK1/RIPK3-mediated necrotic cell death in cardiomyocytes. miR-873 reduces myocardial infarct size upon ischemia/reperfusion (I/R) injury in the animal model. In exploring the molecular mechanism by which miR-873 expression is regulated, we identify NRF as an endogenous sponge RNA and repress miR-873 expression. NRF directly binds to miR-873 and regulates RIPK1/RIPK3 expression and necrosis. Knockdown of NRF antagonizes necrosis in cardiomyocytes and reduces necrosis and myocardial infarction upon I/R injury. Further, we identify that p53 transcriptionally activates NRF expression. P53 regulates cardiomyocytes necrosis and myocardial I/R injury through NRF and miR-873.Our results identify a novel mechanism involving NRF and miR-873 in regulating programmed necrosis in the heart and suggest a potential therapeutic avenue for cardiovascular diseases.
Similar content being viewed by others
Main
Apoptosis has long been thought as the only prototype of programmed cell death and necrosis is traditionally regarded passive and unregulated form of cell death. Emerging evidences indicate that programmed necrosis is a backup cell death program that is activated when apoptotic cell death is blocked.1, 2 It has been demonstrated that necrotic cell death can be tightly regulated by distinct molecules, and the identification of some specific programmed necrotic regulators make it conceivable that necrotic cell death is not only an independent and specialized form of cell death, but that it is also programmed.3, 4, 5 Recent studies have suggested that necrosis is related to various cardiac diseases and is a major contributor to loss of cardiomyocytes cell death.6, 7, 8, 9 However, the molecular mechanisms of programmed necrosis in the heart are unclear relative to apoptosis. New molecules and pathways need to be further discovered.
Death receptors have been shown to induce a particular type of necrotic death in certain cell type, termed programmed necrosis or necroptosis, which is mediated by the receptor-interacting serine/threonine-protein kinase (RIPK) 1 and 3.10, 11, 12 RIPK1 is essential in TNF-α, TRAIL and CD95 ligand-induced necrotic cell death. Upon induction of necrosis, RIPK1 and 3 form a necrosis-inducing complex together.12 RIPK3 phosphorylates the mixed lineage kinase domain-like protein (MLKL) and is a key determinant in mediating the RIPK1 necrotic signaling pathway.13 Inhibition of MLKL blocks the necrotic pathway upon RIPK3 activation and arrest necrosis.13 RIPK1 and RIPK3 have recently been shown to be activated in ischemia heart.5, 14 However, it is not yet clear whether RIPK1 or RIPK3 is a target of microRNAs (miRNAs) and the molecular regulation of RIPK1 or RIPK3 in the programmed necrotic machinery remains to be elucidated in the heart. Addressing these questions will be of great importance to decipher and characterize the molecular pathway of necrosis.
miRNAs are a class of small noncoding RNAs that provide post-transcriptional regulation of gene expression. miRNAs control many cellular processes, including differentiation, proliferation and apoptosis.15, 16, 17, 18, 19, 20 miRNAs also participate in many physiological and pathological processes in cardiovascular health and disease.21, 22, 23 In particular, miRNAs may contribute to the pathophysiological consequences of myocardial infarction, and the alteration of miRNAs levels during apoptosis is reported as a novel therapeutic strategy for cardiac diseases.21, 24, 25 However, it is not yet clear whether miRNAs are involved in the regulation of programmed necrosis by targeting RIPK1/RIPK3 in the heart.
Long noncoding RNAs (LncRNAs) are endogenous regulatory RNAs molecules that do not code for proteins but influence a vast array of biologic processes.26 LncRNAs contain over 200 nucleotides and emerge as new regulators in a wide variety of biological processes, including RNA splicing, epigenetic regulation, cancer generation and development.27, 28, 29, 30 Recent studies suggest critical roles of lncRNAs in mediating the initiation and progression of cardiovascular diseases, and lncRNAs have emerged as novel therapeutic targets in cardiovascular medicine.31, 32, 33, 34 Although lncRNAs play essential roles in various biological processes, hitherto, it is not yet clear whether lncRNAs are involved in the regulation of programmed necrosis in the heart.
Our present work aims at finding out molecules and signaling pathway that are able to regulate cardiomyocyte necrosis. miR-873 was found to participate in the regulation of cardiomyocyte necrosis by targeting RIPK1/RIPK3. miR-873 regulated the expression of RIPK1/RIPK3 by blocking their translation. In searching for the upstream regulator of miR-873, we identified that lncRNA NRF acted as an endogenous sponge that repressed the expression of miR-873. In addition, we demonstrated that NRF can be transcriptionally activated by p53. p53 regulates NRF expression and the consequent cardiomyocyte necrosis through miR-873. The newly identified p53–NRF–miR-873–RIPK1/RIPK3 pathway provides new insight into miR-873 function and lncRNA biology in cardiovascular diseases.
Results
miR-873 participates in the regulation of RIPK1 and RIPK3 expression
It is well known that hydrogen peroxide (H2O2) is an important factor inducing cell death. However, the form of cell death induced by H2O2 varies from apoptosis to necrosis, depending on the exact concentration applied and the cell type tested.35, 36, 37 We tested the effect of H2O2 on cardiomyocytes death and observed that treatment with 0.2 mM H2O2 only triggered apoptosis (Supplementary Figure S1A), while the concentration of 0.6 mM H2O2 preferentially caused necrosis (Supplementary Figure S1B). RIPK1 and RIPK3 are the two critical signaling molecules that mediate the programmed necrosis pathway.10, 11, 12 We firstly tested whether RIPK1 and RIPK3 participated in H2O2-induced cardiomyocytes necrosis. Noticeably, RIPK1 and RIPK3 were dramatically elevated in cardiomyocytes exposed to 600 μM H2O2 (Figure 1a). In the infarcted human hearts, RIPK1 and RIPK3 levels were also markedly increased compared with control hearts (Supplementary Figure S2A). We generated siRNA constructs for RIPK1 and RIPK3, respectively (Supplementary Figures S2B and C) and observed that knockdown of both RIPK1 and RIPK3 attenuated necrotic cell death induced by H2O2 (Figures 1b and c). These data suggest that RIPK1 and RIPK3 are involved in H2O2-induced necrotic cell death in cardiomyocytes.
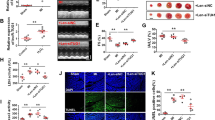
miR-873 participates in the regulation of RIPK1/RIPK3 expression. (a) RIPK1 and RIPK3 expression levels are upregulated upon H2O2 treatment. Cardiomyocytes were treated with 600 μM H2O2 at indicated time. RIPK1 and RIPK3 levels were analyzed by immunoblot (n=3). (b) Knockdown of RIPK1 attenuates necrotic responses induced by H2O2. Cardiomyocytes were infected with adenoviral RIPK1-siRNA or its scramble form (RIPK1-sc), and then treated with 600 μM H2O2. Necrotic cell death was assessed by PI exclusion assay (n=3). *P<0.05 versus H2O2 alone. (c) Knockdown of RIPK3 reduces necrotic responses induced by H2O2. Cardiomyocytes were infected with adenoviral RIPK3-siRNA or its scramble form (RIPK3-sc), and then treated with 600 μM H2O2. Necrotic cell death was assessed by PI assay (n=3). *P<0.05 versus H2O2 alone. (d) Putative miR-873-binding sites in the 3′UTR region of RIPK1 or RIPK3 analyzed by TargetScan program. Mutated miR-873 (miR-873-mut) is shown. (e) miR-873 suppresses the expression of RIPK1 and RIPK3 in cardiomyocytes. Cardiomyocytes were transfected with miR-873 or negative control (NC). RIPK1 and RIPK3 expression levels were analyzed by immunoblot (n=3). (f) Knockdown of miR-873 induces the increase of RIPK1 and RIPK3 expression. Cardiomyocytes were transfected with antagomir miR-873 (anta-873) or its negative control (anta-NC). Forty-eight hours after transfection RIPK1 and RIPK3 expression levels were analyzed by immunoblot (n=3). (g) miR-873 reduces the RIPK1 and RIPK3 expression levels upon H2O2 treatment. Cardiomyocytes were transfected with miR-873 or NC. Twenty-four hours after transfection cells were treated with H2O2. The expression levels of RIPK1 and RIPK3 were analyzed by immunoblot (n=4). (h) Luciferase assay. HEK293 cells were transfected with miR-873 or its mutated form (miR-873-mut), then transfected with the luciferase constructs of the RIPK1 3′UTR or RIPK3 3′UTR. Cells transfected with pGL3 served as a control. The luciferase activity was analyzed (n=3). *P<0.05
To explore the underlying mechanism by which RIPK1 and RIPK3 were upregulated upon H2O2, we tested whether miRNA could control RIPK1 and RIPK3 expression. We analyzed the 3′ untranslated region (UTR) sequence of RIPK1 and RIPK3 by the bioinformatic program TargetScan. We found eight potential miRNAs, each of which had a binding site in the RIPK1 3′UTR and RIPK3 3′UTR. We tested whether these miRNAs could regulate RIPK1 and RIPK3 expression and noticed that only miR-873 resulted in an obvious reduction of RIPK1 and RIPK3 protein levels (Figures 1d and e), whereas other several miRNAs had no effect on the expression of RIPK1 and RIPK3 (Supplementary Figures S3A–G). The inhibitory effect of miR-873 on RIPK1 and RIPK3 expression prompted us to test whether RIPK1 and RIPK3 are downstream targets of miR-873 in controlling necrosis. The mRNA levels of RIPK1 and RIPK3 were not altered upon miR-873 treatment (Supplementary Figures S4A–C). Administration of miR-873 antagomir (Supplementary Figure S4D) induced an elevation of RIPK1 and RIPK3 protein levels (Figure 1f) but not mRNA levels (Supplementary Figures S4E and F). Enforced expression of miR-873 attenuated the increase in RIPK1 and RIPK3 protein levels upon H2O2 treatment (Figure 1g). Thus, it seems that miR-873 regulates RIPK1 and RIPK3 expression at post-transcriptional level. To learn whether RIPK1 and RIPK3 are direct targets of miR-873, we employed the luciferase reporter assay system to test the effect of miR-873 on the translation of RIPK1 and RIPK3. The luciferase reporter assay revealed that miR-873 overexpression induced a decrease in the luciferase activity (Figure 1h). The introduction of mutations in the miR-873 substantially reduced the inhibitory effects of miR-873 (Figures 1d and h). Taken together, it suggests that RIPK1 and RIPK3 are specific targets of miR-873.
miR-873 regulates H2O2-induced necrosis through RIPK1 and RIPK3
Subsequently, we tested whether miR-873 plays an important role in cardiomyocytes necrosis. We firstly detected miR-873 levels and observed that the expression of miR-873 was markedly reduced in the infarcted human hearts (Figure 2a) and in cardiomyocytes exposed to 600 μM H2O2 (Figure 2b). Next, we explored the role of miR-873 in cardiomyocytes necrosis. Electron microscopy was performed to detect the morphological hallmarks of cardiomyocytes. Cardiomyocytes upon exposure to 600 μM H2O2 displayed obvious necrotic morphological characterization such as increase of cell volume, dilation of organelles, rupture of the plasma membrane and loss of cell integrity (Figure 2c). However, enforced expression of miR-873 attenuated the necrotic morphological hallmarks (Figure 2c). In addition, miR-873 also diminished H2O2-induced necrotic cell death, as assessed by propidium iodide (PI) exclusion (Figure 2d). These results indicate an inhibitory effect of miR-873 in cardiomyocytes necrosis. We then explored how miR-873 exerts its effect on necrotic program. Since miR-873 is able to suppress RIPK1/RIPK3 expression, we thus tested whether miR-873 regulated necrosis by targeting RIPK1/RIPK3 in cardiomyocytes. To confirm the relationship between miR-873 and RIPK1/RIPK3 in necrosis program machinery, we employed the target protector technology in which a target protector is able to disrupt the specific interaction of miRNA–mRNA pairs. To this end, we produced RIPK1 and RIPK3 target protector, respectively, and observed that the inhibitory effects of miR-873 on RIPK1/RIPK3 expression and necrotic cell death were significantly abolished in the presence of RIPK1/RIPK3 target protector (Figures 2e and f). These results reveal that miR-873 inhibits necrotic cell death by targeting RIPK1/RIPK3.
miR-873 regulates H2O2-induced necrosis through RIPK1/RIPK3. (a) The levels of miR-873 in human ischemic myocardium. The control non-ischemic human hearts (normal) and ischemic zones of myocardial infarction hearts (infarction) were used for qRT-PCR analysis of miR-873 levels (n=6). *P<0.05 versus normal. (b) The expression levels of miR-873 were analyzed by qRT-PCR in cardiomyocytes exposed to 600 μM H2O2 at the indicated time (n=4). *P<0.05 versus control. (c) Electron microscopy (EM). Cardiomyocytes were transfected with miR-873 or its negative control (NC). Twenty-four hours after transfection cells were treated with 600 μM H2O2. Representative images showed the characteristics of necrotic cells (n=3). (d) miR-873 reduces necrotic responses induced by H2O2. Cardiomyocytes were treated as described in (c). Necrotic cell death was assessed by PI assay. Representative images showed PI-positive cells (left). Red, PI-positive nuclei; blue, DAPI-stained nuclei. Bar=20 μm. The quantitative analysis of necrosis was shown in the right panel (n=4). *P<0.05 versus H2O2 alone. (e and f) RIPK1/RIPK3 target protectors reduce the inhibitory effect of miR-873 on RIPK1/RIPK3 expression and necrosis. Cardiomyocytes were transfected with miR-873, the target protector (RIPK1-TPmiR-873), the target protector (RIPK3-TPmiR-873) or the control (TPcontrol). (e) RIPK1 and RIPK3 expression were analyzed by immunoblot (n=3). (f) Necrotic cell death was assessed by PI assay (n=3). *P<0.05
miR-873 regulates necrotic cell death in the heart
Having demonstrated that miR-873 was involved in H2O2-induced necrosis by targeting RIPK1/RIPK3 in vitro, we further investigated the role of miR-873 and RIPK1/RIPK3 in the pathogenesis of cardiac infarction in the animal model. Mouse model of ischemia/reperfusion (I/R) is widely used for the study of cardiac infarction and I/R has been documented to induce necrosis in the heart.38 miR-873 levels were reduced in response to I/R injury in the ischemic zone but not in the remote zone of heart (Figure 3a). I/R led to an increase in RIPK1/RIPK3 expression, and this increase was attenuated in the presence of miR-873 (Supplementary Figure S4G and Figure 3b). The myosin antibody injection technique was used to assess myocardial necrosis in vivo.38 I/R caused a massive increase in necrotic myosin-positive cells (Figure 3c), whereas the administration of miR-873 resulted in a decrease in the myocardial necrosis (Figure 3c). The administration of miR-873 also reduced myocardial infarct sizes (Figure 3d). Echocardiography on animals revealed that miR-873-administrated mice displayed a decrease in left ventricular internal diameter (Figure 3e) and an amelioration in cardiac function, as indicated by increased fractional shortening (Figure 3f). Thus, our results strongly suggest a possible contribution of miR-873 in inhibiting necrosis in I/R injured heart.
miR-873 regulates necrotic cell death in the heart. (a) miR-873 levels during myocardial ischemia/reperfusion (I/R). Mice were subjected to cardiac I/R at the indicated time. The Sham-operated group served as a negative control. Ischemic zone and Remote zone were prepared at the indicated time for qRT-PCR analysis of miR-873 levels (n=6). *P<0.05 versus 0 min or sham. (b) Enforced expression of miR-873 attenuates the increase of RIPK1/RIPK3 levels in response to I/R jury. Intracoronary delivery of miR-873 and I/R is described in Materials and Methods. RIPK1/RIPK3 levels were analyzed by immunoblot (n=4). (c) miR-873 attenuates myocardial necrosis upon I/R injury. In vivo delivery of miR-873 or NC was performed as described in Materials and Methods. The mice were subjected to sham or I/R as described in the Materials and Methods. Myosin antibody was injected into the mice to label necrotic cells. Representative images of ventricular myocardium sections from sham operation or I/R are shown in the left panel and the quantitative analysis of myosin-positive cells is shown in the right panel. Green, immunohistochemistry for myosin antibody incorporation into the heart; red, wheat germ stain for cell membranes; blue, DAPI-stained nuclei (n=6). Bar=20 μm. *P<0.05 versus WT+I/R. (d) Enforced expression of miR-873 attenuates myocardial infarction sizes. Mice were treated as described in (c). Myocardial infarct sizes were measured as described in the Materials and Methods. The upper panels are myocardial representative photos of midventricular myocardial slices. The lower panel shows infarct sizes. AAR, Area-at-risk; LV, left ventricle; INF, infarct area (n=6). Bar=2 mm. *P<0.05 versus WT+I/R. (e and f) Left ventricular dimensions and cardiac function in mice exposed to I/R were analyzed. Mice were treated as described in (c). Transthoracic echocardiographic analysis was performed after 60 min ischemia followed by 1 week reperfusion. LVIDd, diastolic left ventricular internal diameters; FS, fractional shortening of left ventricular diameter, calculated as [(LVIDd – LVIDs)/LVIDd] × 100. (n=6). *P<0.05 versus WT+I/R
NRF interacts with miR-873 and regulates miR-873 expression
Recent studies have suggested that lncRNAs may act as an endogenous sponge RNA to interact with miRNAs and influence the expression of miRNA.39, 40, 41 To explore the underlying mechanism responsible for miR-873 downregulation in response to H2O2 and I/R treatment, we tested whether lncRNA could participate in the regulation of miR-873 expression. We carried out quantitative reverse transcription polymerase chain reaction (qRT-PCR) to detect lncRNAs levels in response to H2O2 treatment. LncRNAs were chosen from the lncRNA array published online by Fantom company. Among those lncRNAs, five lncRNAs were substantially elevated (Figure 4a). To understand which lncRNA is involved in the regulation of miR-873, we separately knocked down the five lncRNAs by siRNA constructs. The results showed that only knockdown of AK047645 (Supplementary Figure S5A) which we named necrosis-related factor (NRF), led to an increase of miR-873 expression levels (Figure 4b) and the other four lncRNAs had no effect on the miR-873 expression. Enforced expression of NRF (Supplementary Figure S5B) induced a reduction in miR-873 levels (Figure 4c).
NRF interacts with miR-873 and regulates miR-873 expression. (a) LncRNA expression levels upon H2O2 treatment. Cardiomyocytes were untreated (control) or treated with 600 μM H2O2. LncRNAs chosen from Fantom company were analyzed by qRT-PCR (n=3). (b) Knockdown of AK047645 induces the increase of miR-873 expression levels. Cardiomyocytes were infected with indicated adenovirus harboring lncRNA-siRNA (lnc-siRNA) or their scramble form (lnc-sc). Forty-eight hours after infection, miR-873 levels were analyzed by qRT-PCR (n=3). *P<0.05 versus anta-NC. (c) Enforced expression of NRF reduces the expression levels of miR-873. Cardiomyocytes were infected with adenoviral NRF or β-gal. Forty-eight hours after infection miR-873 levels were analyzed by qRT-PCR (n=3). *P<0.05 versus control. (d) NRF contains a site complementary to miR-873. (e) miR-873 can bind directly to NRF in vivo. Cardiomyocytes were transfected with biotinylated wild-type miR-873 (Bio-wt-873) or biotinylated mutant miR-873 (Bio-mut-873). A biotinylated miRNA that is not complementary to NRF was used as a negative control (Bio-NC). Forty-eight hours after transfection, cells were harvested for biotin-based pull-down assay. NRF expression levels were analyzed by real-time PCR (n=3). *P<0.05 versus Bio-NC. (f) NRF can bind to miR-873 in vivo. Cardiomyocytes lysate was incubated with an NRF probe or a random probe-coated magnetic bead. After washing and enrichment of beads/RNA complex, RNA was eluted from the streptavidin beads and was analyzed by northern blot (n=3). I, input (10% samples were loaded); P, pellet (100% samples were loaded). (g) Detection in nuclear or cytoplasmic fractions of NRF and miR-873 in cardiomyocytes. The levels of NRF and miR-873 were analyzed by northern blot (n=3). (h and i) miR-873 could be pulled down by the NRF probe in the cytoplasm (upper panel) but not in the nucleus (low panel). Cells were collected for the analysis of miR-873 by northern blot or by immunoblot with the cellular fractions of cytoplasm (h) or nucleus (i). n=3. I, input (10% samples were loaded); P, pellet (100% samples were loaded). Proliferating cell nuclear antigen (PCNA) is a nuclear marker. Tubulin is a cytoplasmic marker
To understand the mechanism by which NRF regulates the levels of miR-873, we tested whether NRF could interact with miR-873. We compared the sequences of NRF with that of miR-873 using the bioinformatics program RNAhybrid and noticed that NRF contains a binding site of miR-873 (Figure 4d). Further, we applied a biotin–avidin pull-down system to test whether miR-873 could directly bind to NRF. Cardiomyocytes were transfected with biotinylated miR-873, and then harvested for biotin-based pull-down assay. NRF was pulled down by biotinylated wild-type miR-873, but the introduction of mutations that disrupt base pairing between NRF and miR-873 (Supplementary Figure S5C) led to the inability of miR-873 to pull-down NRF (Figure 4e), indicating that the recognition of miR-873 to NRF is sequence specific. We also employed inverse pull-down assay to test if NRF could pull-down miR-873, a biotin-labeled-specific NRF probe was used. The results showed that miR-873 could be co-precipitated by NRF (Figure 4f). Further, we detected the subcellular location of NRF and miR-873. It showed that NRF was expressed both in nucleus and cytoplasm and miR-873 was mainly expressed in the cytoplasm (Figure 4g). So we applied the biotin-labeled-specific NRF probe to assay where the interaction between NRF and miR-873 occurs. The results showed that miR-873 could be co-precipitated by NRF in the cytoplasm (Figure 4h), but not in the nucleus (Figure 4i). Taken together, it appears that NRF is able to directly bind to miR-873 and regulate miR-873 levels.
NRF regulates necrosis through targeting miR-873 and RIPK1/RIPK3
Since NRF can interact with miR-873, we tested whether NRF is able to regulate necrosis through miR-873 and RIPK1/RIPK3. Knockdown of NRF reduced RIPK1/RIPK3 levels (Figure 5a). Overexpression of NRF resulted in the upregulation of RIPK1/RIPK3 expression levels (Figure 5b). NRF counteracted the inhibitory effect of miR-873 on RIPK1/RIPK3 expression (Figure 5c).
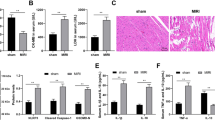
NRF regulates necrosis through targeting miR-873 and RIPK1/RIPK3. (a) Knockdown of NRF reduces the expression levels of RIPK1/RIPK3. Cardiomyocytes were infected with adenoviral NRF-siRNA or NRF-sc. Twenty-four hours after infection RIPK1 and RIPK3 levels were analyzed by immunoblot (n=3). (b) Enforced expression of NRF induces the increase of RIPK1/RIPK3 expression levels. Cardiomyocytes were infected with adenoviral NRF or β-gal. Twenty-four hours after infection RIPK1 and RIPK3 levels were analyzed by immunoblot (n=3). (c) NRF counteracts the inhibitory effect of miR-873 on RIPK1/RIPK3 expression. Cardiomyocytes were transfected with miR-873, and then infected with adenoviral NRF or β-gal. RIPK1 and RIPK3 levels were analyzed by immunoblot (n=4). (d) Knockdown of NRF inhibits H2O2-induced necrotic responses. Cardiomyocytes were infected with adenoviral NRF-siRNA or NRF-sc. Twenty-four hours after infection cells were treated with 600 μM H2O2. Necrosis was assessed by PI assay (n=3). *P<0.05 versus H2O2 alone. (e) NRF levels during myocardial I/R injury. Mice were induced to undergo cardiac I/R. Ischemia zone and remote zone were prepared at the indicated time for qRT-PCR analysis of NRF levels (n=6). *P<0.05 versus 0 min or sham. (f and g) Knockdown of NRF attenuates myocardial necrosis upon I/R injury. Intracoronary delivery of adenoviral constructs of NRF-siRNA or NRF-sc to the hearts was described in the section Materials and Methods. Mice were subjected to I/R as described in the Materials and Methods. Myosin antibody was injected into the mice to label necrotic cells. Representative images of ventricular myocardium sections from sham operation or I/R are shown in (f). Immunohistochemistry for myosin antibody incorporation into the heart (green); cardiomyocytes were identified by α-actinin (red); nuclei are shown in blue. Bar=20 μm. The percentage of cells with myosin antibody infiltration was shown in (g). (n=6). *P<0.05 versus WT+I/R. (h) Knockdown of NRF inhibits myocardial infarction. Mice were treated as described in (f). Infarct size was shown (n=6). AAR, area at risk; LV, left ventricle; INF, infarct area. *P<0.05 versus WT+I/R. (i) Knockdown of NRF preserves cardiac function upon I/R. Transthoracic echocardiographic analysis was performed after 60 min ischemia followed by 1 week reperfusion. Fractional shortening of left ventricular diameter (FS) was shown (n=6). *P<0.05 versus WT+I/R
We then explored the effect of NRF on necrosis. Knockdown of NRF inhibited necrosis induced by H2O2 in cardiomyocytes (Figure 5d). In animal model, NRF levels were increased in response to I/R injury in the ischemic zone (Figure 5e). Knockdown of NRF resulted in a decrease in the myocardial necrosis (Figure 5f and g) and reduced myocardial infarct sizes (Figure 5h). The cardiac function as revealed by fractional shortening was also ameliorated in the NRF knockdown group (Figure 5i). These results indicate that NRF participates in the regulation of cardiomyocytes necrosis and myocardial infarction. Subsequently, we considered whether RIPK1/RIPK3 downregulation by miR-873 contributed to the function of NRF on necrotic cell death. We knocked down the endogenous miR-873 by antagomir, and observed that the inhibitory effect of NRF knockdown on RIPK1/RIPK3 expression (Supplementary Figure S6A) and necrotic cell death (Supplementary Figure S6B) were attenuated in the absence of miR-873. Taken together, these data suggest that NRF mediates necrotic cell death in cardiomyocytes through modulating miR-873-RIPK1/RIPK3 signaling pathway.
p53 regulates NRF expression in transcriptional level
It is reported that mRNAs and miRNAs both can be regulated by transcriptional factors. Thus, we then tested whether NRF also could be regulated at the transcriptional level. We analyzed the promoter region of mouse NRF and observed that there is a potential binding site of p53 (Figure 6a). Then, we tested whether p53 could influence NRF promoter activity. Wild-type NRF promoter (wt) presented a high activity in the presence of p53. And mutations in the p53-binding site (Figure 6a) abolished the promotive effect of p53 on NRF promoter activity (Figure 6b).
NRF is a transcriptional target of p53. (a) Mouse NRF promoter region contains a potential p53-binding site. (b) p53 promotes NRF promoter activity. Cardiomyocytes were treated with the adenoviral β-gal or p53, the constructs of the empty vector (pGL-4.17), the wild-type promoter (wt) or the promoter with mutations in the binding site (mutant), respectively. Luciferase activity was assayed (n=4). *P<0.05. (c) p53 induces the increase of NRF expression levels. Cardiomyocytes were infected with adenoviral β-gal or p53. P53 expression was analyzed by immunoblot (left panel). n=3. NRF levels were analyzed by qRT-PCR (right panel). n=3. *P<0.05 versus control. (d) Knockdown of p53 reduces the NRF expression. Cardiomyocytes were infected with adenoviral p53-siRNA or p53-sc. P53 levels were analyzed by immunoblot (left panel). n=3. NRF levels were analyzed by qRT-PCR (right panel). n=3. *P<0.05 versus control. (e) ChIP-qPCR analysis of p53 binding to the promoter of NRF. Cardiomyocytes were treated with H2O2 at indicated time. Cross-linked chromatin from cardiomyocytes was subjected to immunoprecipitation with antibody against p53 or negative control antibody (lgG). All enrichments were calculated using lgG enrichment as a control in all graphs showing qPCR results. (f) Knockdown of p53 attenuates the increase of NRF promoter activity induced by H2O2. Cardiomyocytes were treated with the adenoviral p53-siRNA or p53-sc, the constructs of the empty vector (pGL-4.17) or the wild-type NRF promoter (wt), and then were treated with H2O2. Luciferase activity was assayed (n=4). *P<0.05
Enforced expression of p53 led to an increase in NRF expression levels (Figure 6c), whereas knockdown of p53 exhibited a decreased expression levels of NRF (Figure 6d), suggesting that NRF may be a potential transcriptional target of p53. Thus, we tested whether p53 could bind to the promoter region of NRF. The chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR) assay revealed that p53 bound to the NRF promoter under the physiological condition. H2O2 treatment enhanced the association of p53 with the NRF promoter (Figure 6e). We further demonstrated that H2O2 induced an elevation in NRF promoter activity in cardiomyocytes (Figure 6f). Concomitantly, knockdown of p53 attenuated the increase of NRF promoter activity upon H2O2 treatment (Figure 6f). These data indicate that NRF can be transcriptionally activated by p53.
p53 regulates necrosis through NRF, miR-873 and RIPK1/RIPK3
We investigated the role of p53 in cardiomyocytes necrosis and myocardial infarction. H2O2 induced an elevation in p53 expression levels (Figure 7a). Knockdown of p53 increased the expression levels of miR-873 (Figure 7b), and attenuated RIPK1/RIPK3 expression (Supplementary Figure S7A) and necrotic cell death (Figure 7c) upon H2O2 treatment in cardiomyocytes. In vivo, p53 levels were also increased in mouse hearts upon I/R injury (Figure 7d) and in the infarcted human hearts (Supplementary Figure S7B). Knockdown of p53 resulted in a decrease in the myocardial necrosis (Figure 7e) and reduced myocardial infarct sizes (Figure 7f). These data indicate that p53 participates in mediating the signal for necrotic cell death in the heart.
p53 regulates necrosis through NRF, miR-873 and RIPK1/RIPK3. (a) p53 levels are increased in cardiomyocytes exposed to H2O2. Cardiomyocytes were exposed to 600 μM H2O2. Cells were harvested at the indicated time for the analysis of p53 levels by immunoblot (n=3). (b) Knockdown of p53 restores the decrease of miR-873 levels induced by H2O2. Cardiomyocytes were infected with adenoviral p53-siRNA or p53-sc. 24 h after infection, cells were treated with H2O2. MiR-873 levels were analyzed by qRT-PCR (n=3). *P<0.05 versus H2O2 alone. (c) Knockdown of p53 reduces necrotic cell death induced by H2O2. Cardiomyocytes were treated as described in (b), PI exclusion was analyzed (n=3). *P<0.05 versus H2O2 alone. (d) The levels of p53 are increased in myocardial I/R injury. Mice were induced to undergo cardiac I/R at indicated time as described in Materials and Methods. P53 levels were analyzed by immunoblot (n=5). (e and f) Knockdown of p53 attenuates myocardial necrosis and myocardial infarction upon I/R injury. Intracoronary delivery of adenoviral constructs of p53-siRNA or p53-sc to the hearts was described in the section of Materials and Methods. Mice were subjected to I/R. Myocardial necrosis (e) and myocardial infarction (f) were analyzed (n=7). *P<0.05 versus WT+I/R. (g and h) Knockdown of miR-873 attenuates the inhibitory effect of p53 knockdown on RIPK1/RIPK3 expression and necrotic cell death induced by H2O2. Cardiomyocytes were infected with adenoviral p53-siRNA, p53-sc, transfected with anta-873 or anta-NC, and then exposed to 600 μM H2O2. RIPK1 and RIPK3 levels were analyzed by immunoblot (g). n=4. Necrotic cells were analyzed by PI assay (h). n=4. *P<0.05
We further tested whether miR-873 and RIPK1/RIPK3 could act as the downstream targets of p53 in necrotic signal pathway. We observed that knockdown of miR-873 attenuated the inhibitory effect of p53 knockdown on RIPK1/RIPK3 expression (Figure 7g) and necrotic cell death (Figure 7h). Thus, it appears that p53 targets NRF, miR-873 and RIPK1/RIPK3 axis in the necrotic cascades.
Discussion
Although the function of miRNAs has been widely studied in cardiomyocytes, the mechanism of miRNAs regulating programmed necrosis in the heart remains largely unknown. miR-873 has been demonstrated to be involved in the pathogenesis of experimental autoimmune encephalomyelitis42 and human T-cell leukemia virus type 1 (HTLV-1).43 But the function of miR-873 in the heart is unclear. Our present study demonstrated that miR-873 participated in the regulation of programmed necrosis and myocardial infarction through targeting RIPK1/RIPK3 in cardiomyocyte. miR-873 has been revealed to be downregulated in colorectal cancer and glioblastoma.44, 45 Whether miR-873 is involved in the regulation of necrosis in other tissues or cell types such as cancer is an interesting question for future investigation. Our study has provided important evidences about the function of miRNAs in the programmed necrosis pathway, a step toward better understanding the miRNA-based therapy for I/R injury and heart failure.
The major breakthrough in the study of programmed necrosis comes from the discovery RIPK146 and RIPK3,10, 12 the enzymatic activity of which are important in the execution of necrosis. RIPK1 and RIPK3 are essential for TNF α-induced programmed necrosis.46, 47 Knockdown of RIPK1 and RIPK3 significantly attenuates TNF-induced necrosis in human and mouse cells.48 The present study demonstrated that RIPK1 and RIPK3 participated in the regulation of programmed necrosis induced by H2O2 in cardiomyocytes. Consistent with previous studies, data from this study further supported the critical role of RIPK1 and RIPK3 in necrotic cell death. In addition, several death receptors have been reported to mediate necrotic cell death upon activation of RIPK1 and RIPK3, such as TNFR1,49 TNFR250 and DR4/5.51 RIP and TRAF2 form a complex upon oxidative stress-induced cell death, but without the participation of TNFR1.50 It remains to be further determined whether the programmed necrosis induced by H2O2 treatment in cardiomyocyte also require some of the signaling molecules such TRAF2/ TNFR1/TNFR2 to convey the cell death signals.
LncRNAs have been defined to play critical roles in diverse physiological and pathological processes.27, 28, 29, 52 Despite the biological importance of lncRNAs, it is not yet clear whether lncRNAs is involved in the regulation of necrosis. The present work reveals a novel function of lncRNA in regulating cardiomyocyte necrosis. Our results showed that knockdown of lncRNA NRF attenuated H2O2-induced necrotic responses in cardiomyocytes and reduced myocardial infracted sizes upon I/R injury. This work provides a new clue for the understanding of lncRNAs-controlled cellular events. The discovery of NRF in necrotic cell death may shed new light on understanding the complex molecular mechanisms of cardiac infarction.
Emerging evidences suggest that lncRNAs may act as endogenous sponge RNAs to interact with miRNAs and influence the expression of miRNA target genes. Hmga2 regulates let-7 activity and promotes lung cancer progression by acting like an endogenous competing RNA for let-7.53 Highly upregulated liver cancer acts as an endogenous ‘sponge’, which downregulates miR-372 leading to reducing translational repression of its target gene, PRKACB.40 Consist with these reports, our results showed that lncRNA NRF served as a miR-873 sponge and reduced the expression of miR-873. We speculate that there may exist some mechanisms that can degrade part of the binding miRNA, which is similar to the function of antagomirs that promote the degradation of miRNA.54, 55, 56 But the exact mechanism is still unclear. It is also an interesting scientific topic and we will focus on that in our future research.
The importance of p53 has been found in all major human cancers. It is a tumor suppressor that enforces normal growth control and genomic stability. Also, p53 acts as a transcription factor and regulates a number of genes that promote apoptosis.57 However, few studies have been focused on the p53 function in cell necrosis. A recent study shows that p53 promotes necrotic cell death by interacting with CypD in mouse embryo fibroblasts.58 Consistent with the previous study, our present work further demonstrated the role of p53 in promoting necrotic cell death. In addition, a couple of studies have revealed that H2O2 caused the alteration of p53 modifications other than its protein level. For example, H2O2 increases p53 acetylation by NAD(+)-dependent deacetylase SIRT1.59 H2O2 can induce p53 acetylation at Lys-305 and Lys-382.60 Exposure of fibroblast to H2O2 induces phosphorylation of p53.61 And H2O2 promotes translocation of p53 from cytosol into the nuclei in the oligodendroglia-like cell line.62 In cardiomyocytes, H2O2 not only can markedly increase the expression level of p53 but also can enhance phosphorylation of p53.63 In our study, the increase in p53 expression level upon H2O2 treatment is the major contributor to the transcription activation of NRF. Modulation of p53 or NRF levels may provide a new approach for tackling necrotic cell death and myocardial infarction. It is of great interesting to study whether modifications of p53, such as acetylation and phosphorylation, are also changed during activation of NRF signaling pathway. And in the future we will exert our effort to elucidate the functions of p53 modifications in necrosis.
Hitherto, knowledge about regulators of programmed necrosis remains glaringly incomplete. Our results provide novel evidences demonstrating that p53–NRF–miR-873–RIPK1/RIPK3 constitutes an axis in regulating cardiomyocyte necrosis. Our results reveal that transcription factors, lncRNAs, miRNAs and protein kinases work in concert and orchestrate a cardiomyocyte necrosis pathway. This pathway defines physiological and pathological roles of programmed necrosis in cardiomyocyte in greater detail. Future studies are needed to elucidate how this pathway is integrated into complex necrotic cascades. Our discovery promotes therapeutic interventions for cardiac diseases based on the knowledge of programmed necrosis in the heart. With time, we will discover more and more de novo and inspiring mechanisms that modify necrotic pathways in the heart.
Materials and Methods
Cardiomyocytes culture and treatment
Cardiomyocytes were isolated from 1- to 2-day-old mice as we described.64 Briefly, after dissection hearts were washed, minced in HEPES-buffered saline solution. Tissues were then dispersed in a series of incubations at 37 °C in HEPES-buffered saline solution containing 1.2 mg/ml pancreatin and 0.14 mg/ml collagenase (Worthington, Lakewood, NJ, USA). After centrifugation cells were re-suspended in Dulbecco’s modified Eagle's medium/F-12 (GIBCO, Grand Island, NY, USA) containing 5% heat-inactivated horse serum, 0.1 mM ascorbate, insulin-transferring-sodium selenite media supplement (Sigma, St Louis, MO, USA), 100 U/ml penicillin, 100 μg/ml streptomycin and 0.1 mM bromodeoxyuridine. The dissociated cells were pre-plated at 37 °C for 1 h. The cells were then diluted to 1 × 106 cells/ml and plated in 10 μg/ml laminin-coated different culture dishes according to the specific experimental requirements. Cells were treated with 200 or 600 μM H2O2 except as otherwise indicated elsewhere. Necrotic cell death was assessed by PI exclusion.
Cell transfection with miR-873 duplexes or antagomirs
The miR-873 duplexes were synthesized by GenePharma Co. Ltd (Shanghai, China). MiR-873 mimic sequence was 5′-GCAGGAACUUGUGAGUCUCCU-3′. MiR-873 mutant sequence was 5′-GACAACCCUUGUGAGUCUCCU-3′. Mimic control sequence was 5′-CAGUACUUUUGUGUAGUACAA-3′. Chemically modified antisense oligonucleotides (antagomirs) were used to inhibit endogenous miR-873 expression. The antagomir sequence was 5′-AGGAGACUCACAAGUUCCUGC-3′. The antagomir control sequence was 5′-CAGUACUUUUGUGUAGUACAA-3′. All the bases were 2′-O-methyl-modified (GenePharma Co. Ltd). Cells were transfected with miRNA duplexes (50 nM) or antagomirs (50 nM) using Lipofectamine 2000 (Invitrogen, GrandIsland, NY, USA) according to the manufacturer’s instructions.
Target protector preparation and transfection
Target protector was designed and named as others and we described.65 In brief, RIPK1-TPmiR-873 sequence is 5′-CTTCAGGAAAGCCTGGAACCTTCCC-3′, RIPK3-TPmiR-873 sequence is 5′-CTTCAGGAAGTGGCAAGGACTGTCT-3′, TPcontrol sequence is 5′-TGACAAATGAGACTCTCTCCTCTCC-3′. They were synthesized by Gene Tools (Philomath, OR, USA), and transfected into the cells using the Endo-Porter kit (Gene Tools) according to the kit’s instructions.
qRT-PCR
Stem–loop qRT-PCR for mature miR-873 was performed as described66 on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Total RNA was extracted using Trizol reagent. After DNAse I (Takara, Japan) treatment, RNA was reverse transcribed with reverse transcriptase (ReverTra Ace, Toyobo). The levels of miR-873 analyzed by qRT-PCR were normalized to that of U6. U6 primers were forward: 5′-GCTTCGGCAGCACATATACTAA-3′; reverse: 5′-AACGCTTCACGAATTTGCGT-3′. qRT-PCR for NRF were performed as we described.64 The sequences of NRF primers were forward: 5′-CAGAAGGCAAACTGCCTCTGTT-3′; reverse: 5′-GTTAAAAGTTGATACCAACAGTG-3′. The results were standardized to control values of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). GAPDH forward primer: 5′-TGTGTCCGTCGTGGATCTGA-3′; reverse: 5′-CCTGCTTCACCACCTTCTTGA-3′. The specificity of the PCR amplification was confirmed by agarose gel electrophoresis.
Immunoblot
Immunoblot was performed as we described.67 The anti-RIPK1, anti-RIPK3 and anti-Tubulin antibodies were from Abcam (GrandIsland, NY, USA). The anti-p53 and anti-PCNA antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology, Inc.
Constructions of adenoviruses harboring mouse NRF, NRF-siRNA, RIPK1-siRNA, RIPK3-siRNA and p53-siRNA
To construct adenovirus encoding NRF, NRF was synthesized by PCR using mouse cDNA as the template. The forward primer was 5′-ATTTTTTGGTTATTTCTGACTAGTAC-3′; the reverse primer was 5′-GAAGTATGTTGTTTATTTTTCTATAA-3′. The PCR fragment was finally cloned into the Adeno-X Expression System (Clontech, Otsu, Japan) according to the manufacturer's instructions. The mouse NRF RNAi target sequence is 5′-GGGACAGAGGCCTGTTTGC-3′. A nonrelated, scrambled RNAi without any other match in the mouse genomic sequence was used as a control (5′-GTAGCCGATGTCGAGTGCG-3′). The mouse RIPK1 target sequence is 5′-CAACCGCGCTGAGTACAAT-3′ and the scramble sequence is 5′-CACAGTAGCTCAGACTACG-3′. The mouse RIPK3 target sequence is 5′-ACACGGCACTCCTTGGTAT-3′ and the scramble sequence is 5′-CTGACTCAGTACTAGCTCG-3′. P53 siRNA sequence is 5′-CACATGACGGAGGTCGTGA-3′; the scramble p53-siRNA sequence is 5′-GACGTATGCATAGTCGTCA-3′. The adenoviruses harboring these RNAi constructs were generated using the pSilencer adeno 1.0-CMV System (Ambion, GrandIsland, NY, USA) according to the Kit’s instructions.
The luciferase constructs of RIPK1 3′UTR and RIPK3 3′UTR
RIPK1 3′UTR was amplified by PCR. The forward primer was 5′-CCAGACAAGGTTTCTCAGTTCTC-3′; the reverse primer was 5′-TCTGAAGCACCTATCTTCAGCAG-3′. RIPK3 3′UTR was amplified by PCR. The forward primer was 5′-TTCAGAGAATCACTGCAAGAG-3′; the reverse primer was 5′-CACAGTTAACATGCTATGTTT-3′. The constructs were sequence verified. 3′UTRs were subcloned into the pGL3 vector (Promega, Madison, WI, USA) immediately downstream of the stop codon of the luciferase gene. Luciferase activity assay was performed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Constructions of mouse NRF promoter and its mutant
The NRF promoter was amplified from mouse genome using PCR. The forward primer was 5′-CAGAGTATGTAGATGGCTGTAG-3′. The reverse primer was 5′-TACACAGCTTAAGTTGTTATCG-3′. The promoter fragment was finally cloned into the vector pGL-4.17 (Promega). The introduction of mutations in the putative p53 binding site was performed with the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) using the wild-type vector as a template. The construct was sequenced to check that only the desired mutations had been introduced.
I/R, preparations of area at risk and histology
Male adult C57BL/6 mice (10 weeks old) were obtained from Institute of Laboratory Animal Science of Chinese Academy of Medical Sciences (Beijing, China). For delivery of miR-873, the mice received on three consecutive days, intravenous injections of miR-873 mimic, or its control at a dose of 30 mg/kg body weight in a small volume (0.2 ml) per injection. For intracoronary delivery of adenoviruses, the mice were anesthetized and ventilated with a HX-300S animal ventilator. The chest was entered through a small left anterior thoracotomy, the pericardial sac was then removed and 2 × 1010 moi adenoviruses of NRF-siRNA or 2 × 1011 moi adenoviruses of p53-siRNA were injected with a catheter from the apex of the left ventricle into the aortic root while the aorta and pulmonary arteries were cross-clamped. The clamp was maintained for 20 s when the heart pumped against a closed system. After removal of air and blood, the chest was then closed and the mice were returned back to cage for recovery. Five days after the injection of adenoviruses, the mice were subjected to I/R surgery.
For I/R injury model, mice were subjected to 60 min ischemia, then 24 h or 1 week reperfusion as described.68 The Sham-operated group experienced the same procedure except the snare was left untied. Evans blue dye was treated as described.68 The areas of infarction (INF), area at risk (AAR) and non-ischemic left ventricle (LV) were assessed with computer-assisted planimetry (NIH Image 1.57) by an observer blinded to the sample identity. The ratio of AAR/LV and INF/AAR was calculated as described.68 All animal experimental procedures were approved by the Animal Ethics Review Committee of QingDao University (ethics approval number 2014 QD-026).
Statistical analysis
Data are expressed as the mean±S.E.M. of at least three independent experiments. We used a one-way analysis of variance for multiple comparisons. A value of P<0.05 was considered significant.
Abbreviations
- lncRNA:
-
long noncoding RNA
- miRNAs:
-
microRNAs
- H2O2:
-
hydrogen peroxide
- I/R:
-
ischemia/reperfusion
- PI:
-
propidium iodide
- RIPK1:
-
receptor-interacting serine/threonine-protein kinase 1
- RIPK3:
-
receptor-interacting serine/threonine-protein kinase 3
- qRT-PCR:
-
quantitative reverse transcription polymerase chain reaction
- TP:
-
target protector
References
Cho YS, Park SY, Shin HS, Chan FK . Physiological consequences of programmed necrosis, an alternative form of cell demise. Mol Cells 2010; 29: 327–332.
Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P . Caspases in cell survival, proliferation and differentiation. Cell Death Differ 2007; 14: 44–55.
Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H et al. Cyclophilin d-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature 2005; 434: 652–658.
Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA et al. Loss of cyclophilin d reveals a critical role for mitochondrial permeability transition in cell death. Nature 2005; 434: 658–662.
Oerlemans MI, Liu J, Arslan F, den Ouden K, van Middelaar BJ, Doevendans PA et al. Inhibition of rip1-dependent necrosis prevents adverse cardiac remodeling after myocardial ischemia-reperfusion in vivo. Basic Res Cardiol 2012; 107: 270.
Kingma JG Jr, Yellon DM . Inability of dimethylthiourea to limit tissue necrosis during acute myocardial infarction in rabbits. Free Radic Biol Med 1992; 12: 263–270.
Lissoni P, Pelizzoni F, Mauri O, Perego M, Pittalis S, Barni S . Enhanced secretion of tumour necrosis factor in patients with myocardial infarction. Eur J Med 1992; 1: 277–280.
Bryant D, Becker L, Richardson J, Shelton J, Franco F, Peshock R et al. Cardiac failure in transgenic mice with myocardial expression of tumor necrosis factor-alpha. Circulation 1998; 97: 1375–1381.
Guerra S, Leri A, Wang X, Finato N, Di Loreto C, Beltrami CA et al. Myocyte death in the failing human heart is gender dependent. Circ Res 1999; 85: 856–866.
Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the rip1-rip3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123.
Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC et al. Rip3, an energy metabolism regulator that switches tnf-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336.
He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to tnf-alpha. Cell 2009; 137: 1100–1111.
Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of rip3 kinase. Cell 2012; 148: 213–227.
Luedde M, Lutz M, Carter N, Sosna J, Jacoby C, Vucur M et al. Rip3, a kinase promoting necroptotic cell death, mediates adverse remodelling after myocardial infarction. Cardiovasc Res 2014; 103: 206–216.
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al. The nuclear rnase iii drosha initiates microrna processing. Nature 2003; 425: 415–419.
Ohtani K, Dimmeler S . Control of cardiovascular differentiation by micrornas. Basic Res Cardiol 2011; 106: 5–11.
Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, Srivastava D . Mir-24 inhibits apoptosis and represses bim in mouse cardiomyocytes. J Exp Med 2011; 208: 549–560.
Wang X, Zhang X, Ren XP, Chen J, Liu H, Yang J et al. Microrna-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury. Circulation 2010; 122: 1308–1318.
Sayed D, He M, Hong C, Gao S, Rane S, Yang Z et al. Microrna-21 is a downstream effector of akt that mediates its antiapoptotic effects via suppression of fas ligand. J Biol Chem 2010; 285: 20281–20290.
Li P . Micrornas in cardiac apoptosis. J Cardiovasc Transl Res 2010; 3: 219–224.
Basson M . Micrornas loom large in the heart. Nat Med 2007; 13: 541.
Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking mirna-1-2. Cell 2007; 129: 303–317.
Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B et al. The muscle-specific microrna mir-1 regulates cardiac arrhythmogenic potential by targeting gja1 and kcnj2. Nat Med 2007; 13: 486–491.
Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW et al. Micrornas in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 2007; 116: 258–267.
Chien KR . Molecular medicine: micrornas and the tell-tale heart. Nature 2007; 447: 389–390.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding rna hotair reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076.
Gong C, Maquat LE . Lncrnas transactivate stau1-mediated mrna decay by duplexing with 3' utrs via alu elements. Nature 2011; 470: 284–288.
Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A et al. An architectural role for a nuclear noncoding rna: Neat1 rna is essential for the structure of paraspeckles. Mol Cell 2009; 33: 717–726.
Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS et al. The melanoma-upregulated long noncoding rna spry4-it1 modulates apoptosis and invasion. Cancer Res 2011; 71: 3852–3862.
Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF et al. A noncoding rna is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci USA 2006; 103: 5781–5786.
Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y et al. Long non-coding rnas as master regulators in cardiovascular diseases. Int J Mol Sci 2015; 16: 23651–23667.
Ounzain S, Burdet F, Ibberson M, Pedrazzini T . Discovery and functional characterization of cardiovascular long noncoding rnas. J Mol Cell Cardiol 2015; 89: 17–26.
Gao W, Wang ZM, Zhu M, Lian XQ, Zhao H, Zhao D et al. Altered long noncoding rna expression profiles in the myocardium of rats with ischemic heart failure. J Cardiovasc Med (Hagerstown, MD) 2015; 16: 473–479.
Pinet F, Bauters C . Potential of non-coding rna as biomarkers in heart failure. Med Sci (Paris) 2015; 31: 770–776.
Gardner AM, Xu FH, Fady C, Jacoby FJ, Duffey DC, Tu Y et al. Apoptotic vs. nonapoptotic cytotoxicity induced by hydrogen peroxide. Free Radic Biol Med 1997; 22: 73–83.
Takeda M, Shirato I, Kobayashi M, Endou H . Hydrogen peroxide induces necrosis, apoptosis, oncosis and apoptotic oncosis of mouse terminal proximal straight tubule cells. Nephron 1999; 81: 234–238.
Troyano A, Sancho P, Fernandez C, de Blas E, Bernardi P, Aller P . The selection between apoptosis and necrosis is differentially regulated in hydrogen peroxide-treated and glutathione-depleted human promonocytic cells. Cell Death Differ 2003; 10: 889–898.
Downey JM . Free radicals and their involvement during long-term myocardial ischemia and reperfusion. Annu Rev Physiol 1990; 52: 487–504.
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M et al. A long noncoding rna controls muscle differentiation by functioning as a competing endogenous rna. Cell 2011; 147: 358–369.
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y et al. Creb up-regulates long non-coding rna, hulc expression through interaction with microrna-372 in liver cancer. Nucleic Acids Res 2010; 38: 5366–5383.
Cazalla D, Yario T, Steitz JA . Down-regulation of a host microrna by a herpesvirus saimiri noncoding rna. Science 2010; 328: 1563–1566.
Liu X, He F, Pang R, Zhao D, Qiu W, Shan K et al. Interleukin-17 (il-17)-induced microrna 873 (mir-873) contributes to the pathogenesis of experimental autoimmune encephalomyelitis by targeting a20 ubiquitin-editing enzyme. J Biol Chem 2014; 289: 28971–28986.
Rahman S, Quann K, Pandya D, Singh S, Khan ZK, Jain P . Htlv-1 tax mediated downregulation of mirnas associated with chromatin remodeling factors in t cells with stably integrated viral promoter. PloS One 2012; 7: e34490.
Skalsky RL, Cullen BR . Reduced expression of brain-enriched micrornas in glioblastomas permits targeted regulation of a cell death gene. PloS One 2011; 6: e24248.
Morgan CP, Bale TL . Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci 2011; 31: 11748–11755.
Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase rip as effector molecule. Nat Immunol 2000; 1: 489–495.
Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W et al. Inhibition of caspases increases the sensitivity of l929 cells to necrosis mediated by tumor necrosis factor. J Exp Med 1998; 187: 1477–1485.
Remijsen Q, Goossens V, Grootjans S, Van den Haute C, Vanlangenakker N, Dondelinger Y et al. Depletion of ripk3 or mlkl blocks tnf-driven necroptosis and switches towards a delayed ripk1 kinase-dependent apoptosis. Cell Death Dis 2014; 5: e1004.
Eum HA, Vallabhaneni R, Wang Y, Loughran PA, Stolz DB, Billiar TR . Characterization of disc formation and tnfr1 translocation to mitochondria in tnf-alpha-treated hepatocytes. Am J Pathol 2011; 179: 1221–1229.
Cusson N, Oikemus S, Kilpatrick ED, Cunningham L, Kelliher M . The death domain kinase rip protects thymocytes from tumor necrosis factor receptor type 2-induced cell death. J Exp Med 2002; 196: 15–26.
Bellail AC, Tse MC, Song JH, Phuphanich S, Olson JJ, Sun SY et al. Dr5-mediated disc controls caspase-8 cleavage and initiation of apoptosis in human glioblastomas. J Cell Mol Med 2010; 14: 1303–1317.
Kanduri C . Kcnq1ot1: a chromatin regulatory rna. Semin Cell Dev Biol 2011; 22: 343–350.
Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM et al. Hmga2 functions as a competing endogenous rna to promote lung cancer progression. Nature 2014; 505: 212–217.
Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M et al. Silencing of micrornas in vivo with 'antagomirs'. Nature 2005; 438: 685–689.
Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G et al. Silencing microrna-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med 2012; 18: 1087–1094.
Horwich MD, Zamore PD . Design and delivery of antisense oligonucleotides to block microrna function in cultured drosophila and human cells. Nat Protoc 2008; 3: 1537–1549.
Wei C-L, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006; 124: 207–219.
Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM . P53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 2012; 149: 1536–1548.
Furukawa A, Tada-Oikawa S, Kawanishi S, Oikawa S . H2o2 accelerates cellular senescence by accumulation of acetylated p53 via decrease in the function of sirt1 by nad+ depletion. Cell Physiol Biochem 2007; 20: 45–54.
Wang Y-H, Tsay Y-G, Tan BC-M, Lo W-Y, Lee S-C . Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J Biol Chem 2003; 278: 25568–25576.
Xie S, Wang Q, Wu H, Cogswell J, Lu L, Jhanwar-Uniyal M et al. Reactive oxygen species-induced phosphorylation of p53 on serine 20 is mediated in part by polo-like kinase-3. J Biol Chem 2001; 276: 36194–36199.
Uberti D, Yavin E, Gil S, Ayasola K-R, Goldfinger N, Rotter V . Hydrogen peroxide induces nuclear translocation of p53 and apoptosis in cells of oligodendroglia origin. Mol Brain Res 1999; 65: 167–175.
Long X, Goldenthal MJ, Marín-García J . Oxidative stress enhances phosphorylation of p53 in neonatal rat cardiomyocytes. Mol Cell Biochem 2007; 303: 167–174.
Tan WQ, Wang K, Lv DY, Li PF . Foxo3a inhibits cardiomyocyte hypertrophy through transactivating catalase. J Biol Chem 2008; 283: 29730–29739.
Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF . Mir-23a functions downstream of nfatc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci USA 2009; 106: 12103–12108.
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT et al. Real-time quantification of micrornas by stem-loop rt-pcr. Nucleic Acids Res 2005; 33: e179.
Li PF, Dietz R, von Harsdorf R . P53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by bcl-2. EMBO J 1999; 18: 6027–6036.
Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP et al. Mir-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med 2011; 17: 71–78.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81522005, 81270160, 81470522) and Beijing Natural Science Foundation (7142103) and Natural Science Foundation of Shandong Province (BS2014SW028).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by V Stambolic
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, K., Liu, F., Liu, CY. et al. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ 23, 1394–1405 (2016). https://doi.org/10.1038/cdd.2016.28
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.28
This article is cited by
-
Cuproptosis: p53-regulated metabolic cell death?
Cell Death & Differentiation (2023)
-
Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets
Functional & Integrative Genomics (2023)
-
The Role of P53 in Myocardial Ischemia-Reperfusion Injury
Cardiovascular Drugs and Therapy (2023)
-
The Effects of MicroRNAs in the Development of Heart Failure
Current Cardiology Reports (2023)
-
LncRNA LINC00461 exacerbates myocardial ischemia–reperfusion injury via microRNA-185-3p/Myd88
Molecular Medicine (2022)