Abstract
EZH2 (the Enhancer of Zeste Homolog 2), as a key epigenetic regulator and EMT inducer, participates in a variety of cancer metastasis. EZH2 stability is regulated by several types of post-translational modifications (PTMs).The long non-coding RNAs (lncRNA) have been implicated to have critical roles in multiple carcinogenesis through a wide range of mechanisms, including modulating the stability of proteins. To date, whether the stability of EZH2 protein is regulated by lncRNAs remains unexplored. Here we report the discovery of ANCR modulating the stability of EZH2, and hence in the invasion and metastasis of breast cancer cells. We determined that ANCR potentiated the CDK1-EZH2 interaction, which then increased the intensity of phosphorylation at Thr-345 and Thr-487 sites of EZH2, facilitating EZH2 ubiquitination and hence its degradation. Moreover, we also uncover ANCR is an important player in breast cancer progression and metastasis mainly through decreasing EZH2 stability. More specifically, we initially found that ANCR level was lower in breast cancer tissues and breast cancer cell lines, in contrast to their normal counterparts. We then demonstrated that knockdown of ANCR induced an EMT program and promoted cell migration and invasion in MCF10A (epithelial cells), whereas ectopic expression of ANCR repressed breast cancer cells migration and invasion. Furthermore, we validated in a nude mouse model that overexpression of ANCR in highly malignant and invasive MDA-MB-231 breast cancer cells significantly reduced the ability of the cells to form tumors and prevented the lung metastasis in vivo. Based on these data, our findings define a new mechanism underlying modulation of EZH2 stability by linking ANCR interaction with EZH2 to promote its phosphorylation that facilitates EZH2 degradation and suppresses breast cancer progression.
Similar content being viewed by others
Main
EZH2 (the Enhancer of Zeste Homolog 2) is one component of the Polycomb Repressive Complex 2 (PRC2), which catalyzes the trimethylation of Lys-27 of histone H3 (H3K27me3) to suppress the transcription of target genes. EZH2 has been reported to have important parts in cancer development and metastasis progression.1, 2 High expression of EZH2 has been detected in many solid tumors, including breast cancer; and the EZH2 level exhibits a negative correlation with the patients' outcome.3, 4 In addition, EZH2 can promote breast tumor initiating cells proliferation.5
Although EZH2 can epigenetically regulate a large number of genes, the regulation of EZH2 itself, especially its stability and enzymatic activity modulating by post-translational modifications (PTMs), remains unclear. It has been reported that retinoblastoma (Rb), SOX4 and miR-101 can regulate EZH2 expression in transcriptional level.6, 7, 8 Also, evidence shows that the EZH2 modulation by PTMs is crucial for its protein stability, as well as to its function in regulation of cancer metastasis.9, 10, 11, 12 The phosphorylation of EZH2 Ser-21 residue by AKT represses its methyltransferase activity.10 In addition, earlier studies indicated that phosphorylation of EZH2 at Thr-345 and Thr-487 residues by cyclin-dependent kinase1 (CDK1) promoted EZH2 degradation via proteasome-mediated pathway.11 Moreover, it was reported that the OGT modified EZH2 with O-GlcNAc at Ser-75 maintained EZH2 protein stability via this O-GlcNAcylation modification.12 Recent data revealed that the EZH2 Lys-348 acetylation was able to repress its phosphorylation at Thr-345 and Thr-487 and increase its stability.9 Besides, they also found that the level of Lys-348 acetylation EZH2 was higher in lung cancer tissues compared with the normal lung tissues and high level of EZH2 Lys-348 acetylation in lung cancer patients predicted a poor outcome.9 Thus, the present data suggest that PTMs of EZH2 can increase the EZH2 accumulation in cancer patients, and the stability of EZH2 may have a key role in cancer development and metastasis.
Over the years, breast cancer has become one of the most common malignancies in women; and the distant metastasis has been reported to be the major cause of mortality in breast cancer patients.13, 14, 15 The epithelial-mesenchymal transition (EMT), a developmental process in which epithelial cells lose their polarity and acquire the migratory properties of mesenchymal cells, is known to be a crucial mechanism contributing to cancer metastasis.16 Increasing evidence implicates that EMT is a pivotal step for tumor infiltration and distant metastasis in a variety of carcinomas.17 It has been reported that EZH2 is a positive regulatory factor of EMT program in breast cancer.6 And EZH2 can bind on CDH1(E-cadherin) promoter to suppress E-cadherin expression and promote cancer cells migration and invasion.18, 19
The long non-coding RNAs (lncRNAs) are RNA transcripts longer than 200 nucleotides with little protein-coding capacity.20, 21 A great deal of emerging research work has revealed that lncRNAs are functional in multiple biological processes, including development, differentiation, cellular senescence and carcinogenesis.22, 23, 24, 25, 26 Evidence implies that the cancer-associated lncRNAs may have either oncogenic or tumor suppression effects, and these lncRNAs have attracted intense research attention as a new class of potential cancer diagnostic biomarkers or therapeutic targets.27, 28 An increasing number of studies have indicated that lncRNAs are important regulators of their binding proteins' stability. For instance, it was reported that lncRNA-LET interacted with NF90 to promote its ubiquitination and subsequent degradation.29 The lincRNA-p21 was shown to bind with HIF-1a and VHL to disrupt the VHL–HIF-1a interaction, which inhibited VHL-mediated HIF-1а ubiquitination and caused HIF-1a accumulation.30 Also, lncRNA FAL1 was uncovered to associated with the epigenetic repressor BMI1 and increased its stability.31 As multiple researches have implicated that EZH2 can bind with some of lncRNAs,32, 33, 34, 35, 36 we were interested in determining if there are any lncRNAs that may interact with EZH2 to modulate its stability.
In an initial attempt to identify lncRNAs that interact with EZH2 to regulate its stability, we have tested several lncRNAs that may potentially bind with EZH2;32, 33, 34, 35, 36, 37 among these, we have focused on one lncRNA termed anti-differentiation ncRNA (ANCR or DANCR), because we find it interacts with EZH2 and decreases EZH2 stability in breast cancer cells in our following research. The ANCR is an 855-nucleotide lncRNA, first reported to be downregulated during differentiation. ANCR is indispensable to enforce the undifferentiated cell state within epidermis.38 It has been reported that ANCR can bind with EZH2 in hFOB1.19 cells.35 In this study, we uncover that ANCR is associated with EZH2 to increase the bind of CDK1 with EZH2 and to promote the phosphorylation at Thr-345 and Thr-487 residues of EZH2, resulting in the ubiquitination and subsequent degradation of EZH2. Experimental data from our study also indicate that ANCR is a repressive regulator of EMT in breast cancer cells. More importantly, we demonstrate that overexpression of ANCR in malignant breast cancer cells effectively represses tumorigenesis and metastasis in vivo in immunodeficiency mice. This has been the first evidence that EZH2 stability is regulated by lncRNA, and ANCR suppresses the EMT and invasion in breast cancer cells in vitro, and that it prevents tumorigenesis and distant metastasis in animals in vivo.
Results
ANCR interacted with EZH2 and decreased its protein level
We first aimed to explore whether the stability of EZH2 is regulated by lncRNAs. In our pre-experiments, we tested several lncRNAs that were reported binding with EZH2.32, 33, 34, 35 Among these lncRNAs, we chose ANCR for further study because we found ANCR had an effect on EZH2 stability. We first validated the association of ANCR with EZH2, by using the RNA immunoprecipitation (RIP) with cell extracts from the MCF10A cells. As the results show, we detected the enrichment of ANCR with the EZH2 (Figure 1a). Next, we performed the RNA pull-down assay by using the Flag-MS2bp-MS2bs-based system, in which the MS2-binding protein (MS2bp) specifically binds RNAs that contain MS2-binding sequences (MS2bs) when they are co-expressed, and proteins bound with RNA will also be pulled down at the same time (Figure 1b). By using this system, we pulled down endogenous proteins associated with ANCR and tested by western blotting with anti-EZH2 antibody. Our RNA pull-down assay results implicate the direct interaction between ANCR and EZH2 (Figure 1b).
ANCR interacted with EZH2 and decreased EZH2 expression in protein level. (a) Relative RIP assays detecting the binding of ANCR with EZH2 in MCF10A cells by quantitative PCR (qPCR). The data are presented as the mean±S.D. (n=3; ***P<0.001, Student’s t-test) (b) Immunoblotting detection of EZH2 after Flag-MS2bp-MS2bs-based pull-down assay. ANTI-FLAG M2 Affinity Gel was used for pull-down assays. (c) Western blots of EZH2 protein in shCtrl, shANCR#1 and shANCR#2 cells. (d) Western blots of EZH2 protein in Vector and ANCR cells. (e) qPCR analysis for EZH2 mRNA in shCtrl, shANCR#1 and shANCR#2 cells. (f) qPCR analysis for EZH2 mRNA in Vector and ANCR cells
An interesting point arising from the experiment was that we found knockdown of ANCR increased EZH2 protein expression in MCF10A, MCF7 and MDA-MB-231 cell lines (Figure 1c), and overexpression of ANCR reduced the EZH2 protein level in MCF10A, MCF7, MDA-MB-231 and HEK293T cell lines (Figure 1d and Supplementary Figure S1a). Nevertheless, either overexpression or knockdown of ANCR had little effect on the transcription level of EZH2 mRNA (Figures 1e and f; Supplementary Figure S1b). Thus, our data demonstrate that ANCR can decrease EZH2 protein level without much affecting its transcription.
The association of ANCR and EZH2 enhanced EZH2 binding with CDK1 and promoted ubiquitin–proteasome degradation
As many lncRNAs are shown to regulate the stability of binding proteins,29, 30, 31 an immediate rationale to this phenomenon is that ANCR may affect the stability of EZH2 protein. To substantiate this assumption, we treated MDA-MB-231 cells expressing ectopic ANCR or control Vector with the proteasome inhibitor MG132 and the protein synthesis inhibitor cycloheximide (CHX). The results showed that the EZH2 was modestly increased in ANCR expressing MDA-MB-231 cells treated with MG132 (Figure 2a), and the same results were obtained in MCF10A and MCF7 cell lines (Supplementary Figure S2). Besides, the half-life of EZH2 was much shorter in ANCR expressing MDA-MB-231 cells upon the addition of CHX for 8 h (Figure 2b). We obtained similar results in MCF7-ANCR and MCF7-Vector cells after CHX treatment (Supplementary Figure S3). We then immunoprecipitated EZH2 protein from ANCR expressing MDA-MB-231 and Vector MDA-MB-231 cells to explore the EZH2 ubiquitination status, and the results revealed that the EZH2 ubiquitination was increased in MDA-MB-231-ANCR cells (Figure 2c). In addition, we also detected an increased EZH2 ubiquitination in MCF10A-ANCR and MCF7-ANCR cells, compared with that in corresponding Vector cells after EZH2 was immunoprecipitated (Supplementary Figure S4). By combining these data, we reached the conclusion that ANCR can attenuate EZH2 stability by promoting its ubiquitination.
ANCR promoted EZH2 ubiquitination and degradation by increasing EZH2 Thr-345 and Thr-487 phosphorylation. (a) The EZH2 protein expression was detected by western blotting in Vector and ANCR cells after treatment with MG132 (10 μM) for 0, 3 and 6 h in MDA-MB-231 cells. (b) The EZH2 expression was detected by western blotting in Vector and ANCR cells treated with CHX (100 μg/ml) in MDA-MB-231 cells. (c) Western blots of endogenous EZH2-associated ubiquitination in Vector and ANCR cells after immunoprecipitation with anti-EZH2 antibody in MDA-MB-231 cells. (d) Western blotting analysis for EZH2-Thr-345 phosphorylation and EZH2-Thr-487 phosphorylation in Vector and ANCR cells with the anti-EZH2-Thr-345-P and anti-EZH2-Thr-487-P antibodies in MDA-MB-231 cells. (e) Western blots of endogenous EZH2-associated CDK1 in Vector and ANCR cells after co-immunoprecipitation assay with anti-EZH2 antibody in MDA-MB-231 cells. (f) Western blots of endogenous EZH2-associated CDK1 in shCtrl and shANCR cells after co-immunoprecipitation assay with anti-EZH2 antibody in MCF10A cells. (g and h) RIP assays detecting the binding of ANCR with EZH2 or CDK1 in MCF10A cells by qPCR. Data are presented as the mean±S.D. (n=3; ns, nonsignificant; ***P<0.001, Student’s t-test)
It has been reported that the CDK1-mediated phosphorylation of EZH2 at Thr-345 and Thr-487 sites is necessary for EZH2 ubiquitination and subsequent degradation by ubiquitin–proteasome pathway.11 A recent study shows that lnc-DC can directly bind with signal transducer and activator of transcription 3 (STAT3) to promote STAT3 phosphorylation on Tyr-705 residue.39 These clues intrigued us to speculate that the ANCR–EZH2 interaction may accelerate its ubiquitination degradation by promoting EZH2 Thr-345 and Thr-487 phosphorylation. To prove this speculation, we performed western blotting using specific phosphorylation antibodies of EZH2 on Thr(P)-345 and Thr(P)-487 sites. The results confirmed that overexpression of ANCR indeed increased the phosphorylation of EZH2 Thr-345 and Thr-487 in MDA-MB-231, MCF10A and MCF7 cells (Figure 2d and Supplementary Figure S5).
Subsequently, in a co-immunoprecipitation experiment conducted in MDA-MB-231 and MCF10A cells, we discovered that ectopic expression of ANCR increased CDK1 interaction with EZH2 (Figure 2e and Supplementary Figure S6). In contrast, results of co-immunoprecipitation with ANCR knockdown in MCF10A cells showed that repression of ANCR was able to attenuate the CDK1 binding with EZH2 (Figure 2f). Furthermore, to determine whether CDK1 also interacts with ANCR, we performed the RIP in shCDK1 and shEZH2 MCF10A cells, and the results revealed that knockdown of CDK1 nearly did not affect the ANCR binding with EZH2, whereas knockdown of EZH2 strikingly decreased the CDK1 binding with ANCR (Figures 2g and h; Supplementary Figure S7). We therefore speculate that EZH2 can directly associate with ANCR whereas CDK1 can not. These findings indicate that the ANCR–EZH2 binding is probably able to facilitate CDK1 interaction with EZH2 to promote phosphorylation of EZH2 at Thr-345 and Thr-487, and finally potentiate EZH2 ubiquitination degradation through ubiquitin–proteasome pathway.
The ANCR level was negatively correlated with human breast cancer
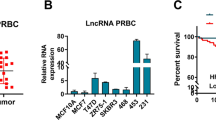
As EZH2 is known to be frequently increased in cancer patients, and higher EZH2 level often lead to the poor prognosis of patients.3, 4, 6 Based on our finding that EZH2 stability was modulated by ANCR, we speculated that ANCR may have a role in breast cancer progression. To determine whether ANCR is linked to breast cancer, we first assessed the ANCR expression in a panel of paired tumor and normal primary tissue specimens that were collected from breast cancer patients (N=32). As can be seen, the qPCR assay revealed that ANCR transcript was expressed at an apparently lower level in tumor tissues compared with the adjacent tissues (Figure 3a). This result indicates that downregulation of ANCR may be common in breast cancer patients. Then we measured the expression of ANCR in breast cancer cell lines, and we found that ANCR level was apparently lower in breast cancer cell lines (MCF7, T47D, BT474, MDA-MB-436, MDA-MB-231 and MDA-MB-231HM) than that in the human normal mammary epithelial cell MCF10A (Figure 3b). These results have substantiated that decreased ANCR expression may be a feature involved in breast tumorigenesis and progression.
ANCR was low expression in clinical breast carcinomas tissues and in breast cancer cell lines. (a) qPCR analysis of ANCR expression in 32 human breast cancerous tissues and non-cancerous tissues. The relative ANCR mRNA level was normalized to β-actin. The statistical differences were analyzed using the t-test. The data are presented as the mean±S.D. (***P<0.001, Student’s t-test) (b) ANCR expression level in breast cancer cell lines and in MCF10A. ANCR mRNA level was normalized to β-actin. The data are presented as the mean±S.D. (n=3; **P<0.01, ***P<0.001, Student’s t-test)
Repression of ANCR induced an EMT program in human mammary epithelial cells
We next intended to explore the biological function of ANCR downregulation in breast carcinoma progression, because a great deal of work has revealed that EZH2 is a key inducer and regulator of EMT program,6, 19 and ANCR is under expressed in breast cancer patients as shown above. We proposed that ANCR may also have effects on EMT program through its regulatory role of EZH2 stability. To validate whether repression of ANCR is able to induce EMT program in MCF10A cells, which expresses high level of endogenous ANCR, we first established stable MCF10A-shANCR cell lines (Figure 4a, left). We then showed that knockdown ANCR in MCF10A cells underwent a change into the spindle-shaped, fibroblastic-like phenotype, with little cell–cell contact (Figure 4c). Besides, the MCF10A-shANCR cells exhibited a significant downregulation of the epithelial marker E-cadherin, accompanied by a prominent upregulation of mesenchymal markers N-cadherin, Vimentin and α-SMA at the mRNA level (Figure 4a). The western blotting results showed that the E-cadherin was decreased, whereas the N-cadherin, Vimentin, Fibronectin and α-SMA were increased in MCF10A-shANCR cells (Figure 4d). The immunofluorescence study further confirmed the results, as we observed a reduced intensity of E-cadherin staining at the cell membrane in MCF10A-shANCR cells. Meanwhile, the Vimentin displayed a weak staining in control cells but intensely stained in MCF10A-shANCR cells (Figure 4e). Furthermore, we detected the endogenous upregulation of a number of other classic EMT inducers, including Twist, Snail, ZEB1 and ZEB2, upon the knockdown of ANCR in MCF10A cells (Figure 4b and Supplementary Figure S8a).
Repression of ANCR induced an EMT program in MCF10A cells. (a) The shRNA-mediated ANCR repression was confirmed by qPCR after lentivirus infection in MCF10A cells (left), and the mRNA expression of EMT markers E-cadherin, N-cadherin, Vimentin and α-SMA was assessed by qPCR. The data are presented as the mean±S.D. (n=3; *P<0.05, **P<0.01, ***P<0.001, Student’s t-test) (b) The expression of EMT inducers (Snail, Twist, ZEB1 and ZEB2) was assessed by qPCR in MCF10A-shANCR and MCF10A-shCtrl cells. The data are presented as the mean±S.D. (n=3; *P<0.05, **P<0.01, Student’s t-test) (c) The morphological change of MCF10A cells after ANCR silencing, as examined by phase contrast microscopy. Scale bar: 100 μm. (d) Western blots of the epithelial marker E-cadherin, and the mesenchymal markers (N-cadherin, Vimentin, Fibronectin and α-SMA) in MCF10A-shANCR and MCF10A-shCtrl cells. (e) Immunofluorescence staining for the EMT markers E-cadherin and Vimentin. Scale bar: 50 μm. (f) Immunoblots of EMT mesenchymal markers Vimentin and α-SMA after ANCR overexpression in MDA-MB-231 cells
We then performed a gain-of-function study in a breast cancer cell line MDA-MB-231, which expresses ANCR at a much lower level. We found that increase of ANCR triggered the down expression of mesenchymal markers (Vimentin and α-SMA), together with the reduced expression of Snail, as shown in the western blots (Figure 4f and Supplementary Figure S8b). Further, we tested whether decreased ANCR also induce EMT in MCF7 breast cancer cells, and the results revealed that ANCR knockdown in MCF7 cells was able to initiate the changes in EMT-related markers (E-cadherin and α-SMA) (Supplementary Figure S9a). Collectively, these data demonstrate that repression of ANCR is sufficient to induce an EMT program.
ANCR facilitated EZH2 target gene expression and inhibited breast cancer cells migration and invasion
A variety of studies demonstrated that EZH2 promoted cancer cells migration through epigenetic repression of many specific tumor suppressor,12, 40 and the EMT program also typically endowed the cells with migratory and invasive capabilities. So we wanted to determine whether repression of ANCR promotes migration and invasion. First, we found that the two EZH2 target genes, HOXA10 and DAB2IP, which are tumor suppressors,41, 42, 43, 44 were decreased in MCF10A-shANCR cells; whereas they were increased upon ectopic expression of ANCR in MDA-MB-231 cells (Figures 5a and b). It has been previously reported that a positive correlation between EZH2 histone methyltransferase (HMT) activity and H3K27me3 exists.45, 46 We demonstrated that repression of ANCR resulted in an increased H3K27me3 global level, whereas ectopic ANCR expression decreased H3K27me3 global level (Supplementary Figure S10). Moreover, increased stability of EZH2 induced by ANCR loss is positively directly correlated with its HMT activity. Furthermore, by using ChIP assays, we revealed that decreased ANCR resulted in much more EZH2 and H3K27me3 enrichments at the promoter regions of EZH2 target gene (E-cadherin, HOXA10 and DAB2IP) (Figures 5c and d). However, overexpression of ANCR decreased the presence of EZH2 and H3K27me3 at these target gene promoters (Supplementary Figure S11). It indicates that ANCR-modulated EZH2 stability can affect EZH2 target gene expression.
Repression of ANCR facilitated the abilities of invasion and migration in breast cancer cells. Experiments in (a–l) were repeated at least three times. Error bars: mean±S.D. (n>3; *P<0.05, **P<0.01, ***P<0.001.Student’s t-test). (a and b) HOXA10 and DAB2IP mRNA expression in MCF10A-shANCR and MDA-MB-231-ANCR cells. (c and d) ChIP assays detecting EZH2 and H3K27me3 enrichments at the EZH2 target gene promoters (E-cadherin, HOXA10 and DAB2IP) in MCF10A-shANCR. (e and f) Western blots of MMP2 and MMP9 proteins in MCF10A-shANCR and MDA-MB-231-ANCR cells. (g and h) Assessment of the MMP2 and MMP9 enzymatic activities by gelatin zymography in MCF10A-shANCR and MDA-MB-231-ANCR cells. (i and j) Migration assays in MCF10A-shANCR#1, MCF10A-shANCR#2 and MDA-MB-231-ANCR cells. (k and l) Invasion assays in MCF10A-shANCR#1, MCF10A-shANCR#2 and MDA-MB-231-ANCR cells
Then, we detected both MMP2 and MMP9 mRNA and protein levels were elevated in MCF10A-shANCR cells compared with that in MCF10A-shCtrl cells (Supplementary Figure S12a and Figure 5e). Gelatin zymography assays further showed that repression of ANCR increased the mature MMP9 activities (Figure 5g). Subsequently, we observed that decreased expression of ANCR significantly accelerated the speed of wound closure in MCF10A cells (Supplementary Figure S13a). Also, we proved that the abilities of migration and invasion in MCF10A-shANCR cells were elevated as revealed by the transwell migration and matrigel invasion assays (Figures 5i and k). In contrast, overexpression of ANCR in MDA-MB-231-ANCR cells downregulated MMP2 and MMP9 expression at both mRNA and protein levels (Supplementary Figure S12b and Figure 5f), and inhibited the MMP9 enzyme activity as revealed by gelatin zymography assay (Figure 5h). Also, the wound healing assay showed that ectopic expression of ANCR repressed wound closure in MDA-MB-231 cells (Supplementary Figure S13b). Besides, overexpression of ANCR in the invasive MDA-MB-231 cells reduced the migratory and invasive behaviors (Figures 5j and l). In addition, we showed that depletion of ANCR increased the motility of MCF7 cells (Supplementary Figures S9b and c). Together, we have proven in the above experiments that ANCR is effective as a negative regulator of EMT and cell migration in human mammary epithelial MCF10A cells, in non-metastatic MCF7 breast cancer cells and in highly metastatic MDA-MB-231 breast cancer cells.
EZH2 was required for ANCR inhibited EMT and suppressed breast cancer cells migration and invasion
To assess whether the EZH2 stability is a direct and main mechanism involved in modulation of knockdown ANCR-induced EMT program and migration, we concomitantly expressed EZH2 in MCF10A-ANCR and MDA-MB-231-ANCR cells. Notably, the forced expression of EZH2 increased the mesenchymal marker Vimentin expression that was repressed by ANCR in MCF10A and MDA-MB-231 cells (Figures 6a and b). Meanwhile, the epithelial maker E-cadherin was decreased in MCF10A-ANCR-EZH2 cells compared with that in MCF10A-ANCR cells (Figure 6a). Moreover, wound healing assays showed that overexpression of EZH2 apparently increased migration speed of both MCF10A-ANCR and MDA-MB-231-ANCR cells (Figure 6c and Supplementary Figure S14). In addition, the transwell migration and matrigel invasion assays further confirmed that ectopic expression of EZH2 enhanced the migration and invasion capabilities of MCF10A-ANCR and MDA-MB-231-ANCR cells (Figures 6d–g). These results clearly indicate that the ANCR-mediated EZH2 degradation is probably critical in attenuation of the ability of breast cancer cells to initiate EMT and metastasis program.
Forced expression of EZH2 restored EMT program and increased migration and invasion in ANCR-overexpressing cells. (a) Western blots of the expression of EZH2 and EMT markers (E-cadherin and Vimentin) in MCF10A-ANCR and MCF10A-ANCR-EZH2 cells. (b) Western blots of the expression of EZH2 and EMT marker Vimentin in MDA-MB-231-ANCR and MDA-MB-231-ANCR-EZH2 cells. (c) Representative images from wound healing assay in MCF10A-ANCR and MCF10A-ANCR-EZH2. Scale bars: 200 μm. (d and e) Migration assays in MCF10A-ANCR-EZH2 and MDA-MB-231-ANCR-EZH2 cells. The data are presented as the mean±S.D. (n=3; **P<0.01, ***P<0.001,Student’s t-test). (f and g) Invasion assays in MCF10A-ANCR-EZH2 and MDA-MB-231-ANCR-EZH2 cells. The data are presented as the mean±S.D. (n=3; **P<0.01, ***P<0.001,Student’s t-test)
ANCR repressed breast cancer tumorigenesis and metastasis in vivo
Next, we intended to evaluate the physiological and pathological relevance of ANCR to the tumorigenesis and metastasis in breast cancer in vivo. BABL/c nude mice were subcutaneously injected with either MDA-MB-231-Vector or MDA-MB-231-ANCR cells. The results showed that mice injected with MDA-MB-231-Vector cells formed apparently larger tumors than that injected with MDA-MB-231-ANCR cells (Figures 7a–e), indicating that ANCR was able to repress tumor growth in vivo. Furthermore, to determine whether ANCR can affect breast cancer cell metastasis, the MDA-MB-231-Vector or MDA-MB-231-ANCR cells were injected into the tail veins of nude mice. Strikingly, we discovered that MDA-MB-231-ANCR cells metastasized to the lungs of nude mice less effectively, in contrast to the MDA-MB-231-Vector cells, as illustrated by the bioluminescence imaging (Figures 7f and g). Noticeably, histological examination revealed that all mice bearing MDA-MB-231-Vector cells had a larger number of macroscopic lung metastasis compared with mice transplanted with MDA-MB-231-ANCR cells (Figures 7h and i). Subsequently, the lung tissue sections were prepared and examined after stained with hematoxylin and eosin, and we detected smaller and fewer metastatic foci in the tissues from mice injected with MDA-MB-231-ANCR cells (Figure 7j). Finally, we measured the EZH2 level in lung tissue sections by using the immunohistochemistry, and we detected a reduced EZH2 level in mice injected with MDA-MB-231-ANCR cells (Figure 7k). Together, these data strongly suggest that ANCR has a pivotal role in repression of breast cancer tumorigenesis and metastasis in vivo.
ANCR repressed tumorigenesis and metastasis in vivo. (a–e) Representative photographs (a) and bioluminescence images (b) of tumorigenesis via subcutaneous injection of Vector or ANCR MDA-MB-231 cells into BALB/c female nude mice (N=8, for each experimental group). After 9 weeks the xenograft tumors were detached (e), weighted (c) and measured for volume (d). The tumor weights and volumes are presented as the means±S.D. (**P<0.01, ***P<0.001, Student’s t-test). (f and g) Representative bioluminescence images of lung metastasis (f) in mice injected with cells as indicated via tail veins, and the metastasis were quantified by measuring the photo flux (g). (h–j) After 9 weeks, nude mice were sacrificed and lung metastatic nodules were examined macroscopically or detected by H&E staining. The white arrows denote the metastatic nodules. Error bars in h represent mean±S.D. (***P<0.001, Student’s t-test). (k) Representative images of the immunohistochemical staining of EZH2 in nude mice lung metastasis sections. Scale bars: 100 μm
Discussion
Data provided in this report identified a novel regulatory mechanism of EZH2 PTMs by ANCR. Our results indicate that ANCR interacts with EZH2 and facilitates CDK1 binding with EZH2 to promote its phosphorylation at Thr-345 and Thr-487 sites, resulting in EZH2 degradation. Besides, we also reveal that ANCR is a critical negative regulator of EMT program mainly through affecting the EZH2 stability, and it inhibits breast cancer cells migration and invasion in vitro and in vivo.
Evidence from this and other studies demonstrate that EZH2 phosphorylation is an important regulatory mechanism of its functions and its interactions with other proteins. It was reported that phosphorylation of EZH2 at serine 21 by AKT signaling pathway decreased lysine 27 trimethylation and de-repressed the silenced target genes.10 A recent study also suggests that EZH2 phosphorylation at Serine 21 enhances STAT3 methylation and activation in glioblastoma multiforme stem-like cells.47 Moreover, phosphorylation of EZH2 at Thr-345 and Thr-487 residues by CDK1 results in reduced EZH2 stability.11 Besides, phosphorylation at Thr-345 and Thr-487 of EZH2 by CDK1 is also critical for the EZH2-mediated epigenetic gene silencing in breast and prostate cancer cells.48, 49 It is known that phosphorylation at sites Thr-345 and Thr-487 of EZH2 executes opposite role in mediating gene silencing in prostate and breast cancer cells.48, 49 Findings in our study indicate that ANCR mediated the instability of EZH2 by enhancing the level of phosphorylation at both Thr-345 and Thr-487 in breast cancer cells, with a concomitant inhibition of breast cancer cell motility. We also revealed that knockdown of CDK1 increased EZH2 protein level. Significantly, by using the CDK1 specific inhibitor CGP74514A (CGP), we demonstrated that the phosphorylation of EZH2 at Thr-345 and Thr-487 of was predominantly repressed, and in the meantime the EZH2 protein level was increased upon the addition of the CGP in MCF10A-ANCR cells (Supplementary Figure S15). These experiments suggest that the ANCR-mediated EZH2 instability requires the CDK1 kinase activity.
A recent study proposes that the lncRNA lnc-DC directly interacts with STAT3 through its 3′-end stem-loop structure, and promotes the phosphorylation on Tyr-705 of STAT3 by preventing STAT3 binding to SHP1 for dephosphorylation.39 At the present time, the details of how ANCR physically interacts with EZH2 and how this interaction promotes CDK1 binding with EZH2 still remain unclear. Presumably, ANCR may associate with EZH2 in a similar way to that of lnc-DC, that is, ANCR interacts with EZH2 by stem-loop structure to change the conformation of EZH2, which probably facilitates the recognition and binding of CDK1 on EZH2 to phosphorylate and finally degrades the protein through the ubiquitin–proteasome pathway (Supplementary Figure S16). This hypothesis needs further investigations based on bioinformatic and structural analyses. Despite this, it appears that interaction with target proteins to influence their PTMs, especially the phosphorylation, may be a major functional mechanism of lncRNAs.
Interestingly, a report that has just come out shows that ANCR (termed DANCR in that report) increases the stemness features of hepatocellular carcinoma cells to promote tumorigenesis and hepatic tumor colonization by increasing the β-catenin mRNA stabilization.50 Meanwhile, recent investigations also suggest that ANCR can repress Wnt/β-catenin signaling pathway during differentiation.37, 51 Their data suggest that knockdown of ANCR promotes the proliferation of periodontal ligament stem cells (PDLSCs), and the β-catenin mRNA level is much higher in PDLSC-shANCR cells.37 Also, ANCR suppresses odontoblast-like differentiation by inhibiting the Wnt/β-catenin signaling pathway, and upregulation of ANCR decreases the p-GSK-3β and β-catenin expression in human dental pulp cells.51 We indicate in this study that depletion of ANCR promotes cell proliferation in MCF10A cells; whereas ectopic expression of ANCR represses the proliferation ability of MDA-MB-231 cells and MCF10A cells (Supplementary Figure S17). These studies suggest a variable ANCR expression pattern and biological function in different cancer types and different cell lines.
We further analyzed the correlation between ANCR and the breast cancer subtypes, and we found that ANCR expression was lower in ER− breast cancer tissues compared with that in ER+ counterparts. Also, the ANCR level was much lower in Her2− compared with Her2+ breast cancer tissues. Besides, we revealed that ANCR was expressed in a reduced level in metastasis breast cancer tissues. Nevertheless, we did not detect the correlation between ANCR and PR expression (Supplementary Figure S18). As inhibition of EZH2 has increasingly shown its clinic value in cancer prevention,1 it would become logical that ANCR may be a potential biomarker or therapeutic target for breast cancers.
Overall, this study discovers that the EZH2 stability is able to modulated by ANCR, and ANCR is a novel potential tumor suppressor in breast cancer through modulating the EMT process and cells migration. More importantly, the conclusion that overexpression of ANCR in aggressive breast cancer cells dramatically blocked tumor growth and metastasis in vivo, points to the possibility for the development of ANCR as a potent target of therapeutic interventions for highly aggressive and malignant breast cancers.
Materials and Methods
Specimens and cell lines
Breast tissue specimens were obtained from the Oncology Hospital of Jilin Province, China. Samples were frozen in liquid nitrogen immediately after surgery. All breast tissues were collected using the protocols approved by the Ethics Committee of the Jilin Oncology Hospital. All the cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). And the detail of cell lines and cells culture are shown in Supplementary Information.
Antibodies and plasmids
Antibodies and plasmids have been shown in Supplementary Information.
Immunofluorescence, immunoprecipitation
Immunofluorescence and Immunoprecipitation was performed as described previously.52
RNA extraction, reverse transcription and quantitative RT-PCR
Experiments were carried out according to the protocol practically used in this laboratory.53 The sequences of PCR primers are shown in Supplementary Information.
Wound healing, cell invasion, migration and proliferation assays
Experiments for determination of cell invasion, migration and proliferation were performed as described previously.54 For wound healing assay, 5 × 105 cells were plated in 6-well plates. The scratch was done by a 10 μl pipette tip. The progression of migration was detected and photographed after 24 h.
RIP assay and RNA pull-down assay
The RIP experiment was carried out using the EZ-Magna RIP Kit (Millipore, Billerica, MA, USA Catalog No. 17-701) according to the manufacturer's protocol with 5 μg of anti-EZH2 antibody.
The co-precipitated RNAs were extracted through Trizol reagent (Invitrogen, Grand Island, NY, USA, Catalog No.15596026) and detected by RT-PCR or real-time PCR.
The Flag-MS2bp-MS2bs-based RNA pull-down assay was carried out by using the ANTI-FLAG M2 Affinity Gel (Sigma, St Louis, MO, USA, Catalog Number A2220). And the RNA pull-down assay details are shown in Supplementary Information.
In vivo tumor formation and lung-colonization assays
In vivo tumor formation and lung-colonization assays are shown in Supplementary Information.
Statistical analysis
Data are presented as mean±S.D. The Student t-test (2-tailed) was used to determine statistic significance of differences between groups. P<0.05 was considered statistically significant. Statistical analysis was performed using the GraphPad Prism software (GraphPad Software, La Jolla, CA, USA).
Abbreviations
- EZH2:
-
enhancer of zeste homolog 2
- PTMs:
-
post-translational modifications
- lncRNAs:
-
long non-coding RNAs
- CDK1:
-
cyclin-dependent kinase1
- ANCR:
-
anti-differentiation ncRNA
- EMT:
-
epithelial-mesenchymal transition
- α-SMA:
-
α-smooth muscle actin
- FN:
-
fibronectin
- MMP2:
-
matrix metalloproteinase 2
- MMP9:
-
matrix metalloproteinase 9
- H3K27me3:
-
trimethylation of Lys-27 of histone H3
- ER:
-
estrogen receptor alpha
- PR:
-
progesterone receptor
- Her2 or ERBB2:
-
human epidermal growth factor receptor 2
References
Kim KH, Roberts CW . Targeting EZH2 in cancer. Nat Med 2016; 22: 128–134.
Simon JA, Lange CA . Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 2008; 647: 21–29.
Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 2003; 100: 11606–11611.
Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629.
Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell 2011; 19: 86–100.
Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 2013; 23: 768–783.
Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K . EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22: 5323–5335.
Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008; 322: 1695–1699.
Wan J, Zhan J, Li S, Ma J, Xu W, Liu C et al. PCAF-primed EZH2 acetylation regulates its stability and promotes lung adenocarcinoma progression. Nucleic Acids Res 2015; 43: 3591–3604.
Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 2005; 310: 306–310.
Wu SC, Zhang Y . Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem 2011; 286: 28511–28519.
Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC et al. O-GlcNAcylationregulates EZH2 protein stability and function. Proc Natl Acad Sci USA 2014; 111: 1355–1360.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM . Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Gupta GP, Massague J . Cancer metastasis: building a framework. Cell 2006; 127: 679–695.
Thiery JP . Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2: 442–454.
Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA . Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest 2009; 119: 1438–1449.
Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008; 27: 7274–7284.
Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene 2012; 31: 583–594.
Guttman M, Rinn JL . Modular regulatory principles of large non-coding RNAs. Nature 2012; 482: 339–346.
Frith MC, Bailey TL, Kasukawa T, Mignone F, Kummerfeld SK, Madera M et al. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol 2006; 3: 40–48.
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477: 295–300.
Ponting CP, Oliver PL, Reik W . Evolution and functions of long noncoding RNAs. Cell 2009; 136: 629–641.
Puvvula PK, Desetty RD, Pineau P, Marchio A, Moon A, Dejean A et al. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat Commun 2014; 5: 5323.
Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol 2013; 20: 908–913.
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ 2013; 20: 1558–1568.
Li Q, Su Z, Xu X, Liu G, Song X, Wang R et al. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc Natl Acad Sci USA 2012; 109: 14110–14115.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076.
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 2013; 49: 1083–1096.
Yang F, Zhang H, Mei Y, Wu M . Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell 2014; 53: 88–100.
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 2014; 26: 344–357.
Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–693.
Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and Interacts with miR-205. Cancer Res 2015; 75: 1322–1331.
Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y et al. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget 2015; 6: 32410–32425.
Zhu L, Xu PC . Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun 2013; 432: 612–617.
Benetatos L, Voulgaris E, Vartholomatos G, Hatzimichael E . Non-coding RNAs and EZH2 interactions in cancer: long and short tales from the transcriptome. Int J Cancer 2013; 133: 267–274.
Jia Q, Jiang W, Ni L . Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol 2015; 60: 234–241.
Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 2012; 26: 338–343.
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014; 344: 310–313.
Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun 2015; 6: 6051.
Yoshida H, Broaddus R, Cheng W, Xie S, Naora H . Deregulation of the HOXA10 homeobox gene in endometrial carcinoma: role in epithelial-mesenchymal transition. Cancer Res 2006; 66: 889–897.
Smits M, van Rijn S, Hulleman E, Biesmans D, van Vuurden DG, Kool M et al. EZH2-regulated DAB2IP is a medulloblastoma tumor suppressor and a positive marker for survival. Clin Cancer Res 2012; 18: 4048–4058.
Wang J, Zhu X, Hu J, He G, Li X, Wu P et al. The positive feedback between Snail and DAB2IP regulates EMT, invasion and metastasis in colorectal cancer. Oncotarget 2015; 6: 27427–27439.
Xie D, Gore C, Liu J, Pong RC, Mason R, Hao G et al. Role of DAB2IP in modulating epithelial-to-mesenchymal transition and prostate cancer metastasis. Proc Natl Acad Sci USA 2010; 107: 2485–2490.
Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 2008; 10: 1291–1300.
Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 2015; 21: 1491–1496.
Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013; 23: 839–852.
Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 2011; 13: 87–94.
Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol 2010; 12: 1108–1114.
Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2015; 63: 499–511.
Chen L, Song Z, Huang S, Wang R, Qin W, Guo J et al. lncRNA DANCR suppresses odontoblast-like differentiation of human dental pulp cells by inhibiting wnt/beta-catenin pathway. Cell Tissue Res 2015; 364: 309–318.
Zhao L, Zhang Y, Gao Y, Geng P, Lu Y, Liu X et al. JMJD3 promotes SAHF formation in senescent WI38 cells by triggering an interplay between demethylation and phosphorylation of RB protein. Cell Death Differ 2015; 22: 1630–1640.
Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 2014; 5: e1287.
Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res 2012; 72: 4597–4608.
Acknowledgements
We thank Dr. Xiaofei Zheng (Academy of Military Medical Sciences, China) for providing the plasmids mentioned in Supplementary Information. This work was supported by the grants from the National Natural Science Foundation of China (grant numbers: 31571317, 31570718, 31571478, 31371294, 31170719 and 31271442).
Author contributions
Conception and design by Zhongwei Li, Jun Lu, Baiqu Huang; Development of methodology by Zhongwei Li; Acquisition of data by Zhongwei Li, Pingfu Hou, Dongmei Fan, Meichen Dong, Ruosi Yao, Pengyu Geng, Adhanom Mihretab; Computational analysis by Zhongwei Li; Writing by Baiqu Huang, Ju Lu, Zhongwei Li. Administrative, technical or material support by Hongyuan Li, Musong Ma, Guannan Wang, Yuxin Li; Study supervision by Yu Zhang, Jun Lu, Baiqu Huang.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by R De Maria
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Z., Hou, P., Fan, D. et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ 24, 59–71 (2017). https://doi.org/10.1038/cdd.2016.95
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.95
This article is cited by
-
FBXO22 promotes glioblastoma malignant progression by mediating VHL ubiquitination and degradation
Cell Death Discovery (2024)
-
Assessing the expression of differentiation antagonizing non-protein coding RNA (DANCR) in newly diagnosed Egyptian acute myeloid leukemia patients
The Egyptian Journal of Internal Medicine (2023)
-
LINC00543 promotes colorectal cancer metastasis by driving EMT and inducing the M2 polarization of tumor associated macrophages
Journal of Translational Medicine (2023)
-
RETRACTED ARTICLE: LncRNA BCAR4 promotes migration, invasion, and chemo-resistance by inhibiting miR-644a in breast cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
TRIM21 attenuates renal carcinoma lipogenesis and malignancy by regulating SREBF1 protein stability
Journal of Experimental & Clinical Cancer Research (2023)