Abstract
Caspases are aspartate-specific cysteine proteases that have an essential role in apoptosis and inflammation, and contribute to the maintenance of homeostasis in the intestine. These facts, together with the knowledge that caspases are implicated in host-microbe crosstalk, prompted us to investigate the effect of caspase (Casp)1, -3 and -7 deficiency on the composition of the murine gut microbiota. We observed significant changes in the abundance of the Firmicutes and Bacteroidetes phyla, in particular the Lachnospiraceae, Porphyromonodaceae and Prevotellacea families, when comparing Casp-1, -7 and -3 knockout mice with wild-type mice. Our data point toward an intricate relationship between these caspases and the composition of the murine gut microflora.
Similar content being viewed by others
Main
Continuous apoptosis and shedding of intestinal epithelial cells is a unique feature of the intestine, and tight regulation of cell death is essential for maintaining the intestinal barrier. Damage to the thin epithelial cell layer can result in invasion of bacteria into the lamina propria and the subsequent activation of immune cells residing there. Because of their key roles in apoptosis and inflammation,1 the apoptotic executioner caspase (Casp)3 and -7, and the inflammatory Casp-1, all serve important functions in preservation of the barrier and maintenance of homeostasis in the intestine. Aberrant inflammatory and apoptotic caspase activity has been described in a large number of human diseases, including inflammatory bowel disease and colorectal cancer.
Casp-3 and Casp-7 exhibit almost identical specificity toward certain synthetic peptide substrates,2 and their functions in the cell death machinery are generally thought to be redundant. However, evidence is accumulating that they have distinct roles in apoptosis and mediate a larger number of non-apoptotic functions than previously expected, including a role in inflammation. Inflammatory and apoptotic caspases might modulate innate immunity in opposing ways. For example, the inflammatory Casp-1 proteolytically activates the pro-inflammatory cytokines interleukin (IL)-1 and IL-18, whereas Casp-3 and -7 proteolytically inactivate IL-33.3 IL-33 functions as an alarmin during tissue damage or inflammation by its effects on both innate and acquired immunity. Interestingly, IL-33 was recently found to be increased in ulcerative colitis.4
The involvement of Casp-1 in intestinal pathology has been extensively studied. Engagement of pattern recognition receptors by specific pathogen-associated molecular patterns leads to the activation of nuclear factor kappa B, microtubule-associated protein kinase and inflammasomes (in turn activating Casp-1), as well as interferon responses.5 This activation cascade eventually results in an innate and adaptive immune response. In vivo experimental models of infection have demonstrated that Casp-1-knockout (KO) mice are more susceptible to a variety of infections.6 The role of Casp-1 in dextran sulfate sodium (DSS)-induced colitis is controversial. Some papers reported that KO of Casp-1 protects mice against DSS colitis, whereas others reported that it sensitizes them for it.7, 8, 9 In line with this reported sensitization, DSS causes a more severe colitis in mice deficient in other components of the inflammasome protein complex, for example, apoptosis-associated speck-like protein containing a CARD, NRLP3 and NRLP6.8, 10
Several studies probed the impact of the intestinal bacteria on host gene and protein expression, and ample evidence exists that bacteria have an effect on caspase activation and vice versa.11, 12 As apoptosis can act as an innate defense strategy, it is likely that aberrant apoptosis affects the microbial composition of the host. For example, apoptosis of macrophages infected with M. tuberculosis is associated with diminished pathogen viability.13 The delayed apoptosis of neutrophils after bacterial infection is another defense mechanism; it lengthens the lifespan of these cells to ensure effective clearance of bacteria.14 A third example is the release of antimicrobial proteins, such as histone H1, by apoptotic intestinal epithelial cells to prevent invasion by commensal and pathogenic bacteria.15
In most published studies, it was assumed that aberrant caspase expression or activation in host cells affects the immune system and/or cell death machinery, the host alone being responsible for the manifested phenotype. However, experiments on germ-free mice and treatment with antibiotics already pointed out the important role of the microbiome in murine colitis and other diseases.16 Interestingly, a recent study linked the absence of the inflammasome component NRLP3 to a change in gut microbiota composition in naive mice.10 Today, the link between host (patho)physiology and the gut microbiome is being increasingly recognized and studied.10, 17, 18 In-depth knowledge of the bacterial ecosystem in relation to disease might pave the way for the generation of customized therapeutics and probiotics.
The host microbiota can be considered as a complex trait that is under host genetic control.19, 20 Mouse models are crucial for the study of single-gene changes in the genome and are used widely to gain insight into the biology of human diseases. We examined whether the absence of Casp-1, -3 and -7 affects the composition of the gut microbiota of naive mice. To test this hypothesis, we analyzed the gut microbiome of Casp-1, -3 and -7-deficient mice using high-throughput 16S rRNA gene pyrosequencing.
Results and Discussion
General characteristics of the microbiomes of Casp-KO and -wild-type (WT) mice
On a C57/BL6 background, mice lacking Casp-3 or -7 have a normal life span and display a limited apoptotic phenotype.21 Pro- and anti-inflammatory cytokine levels in the blood of these mice do not differ in unchallenged conditions, and are similar to levels in WT mice (data not shown). Casp-1-deficient mice are also overtly normal.22 Examination of the intestines of Casp-1, -3 and -7-deficient mice shows that they do not differ morphologically from the WT intestine as determined i.a. by hematoxylin and eosin staining of intestinal tissue sections and colon length (data not shown). Because we were interested in establishing the physiological effects of caspase deficiency on the intestinal microbiota, this study was performed with naive mice. DNA was extracted from the cecal contents of five mice of each genotype, bacterial 16S rRNA genes were amplified by PCR, and the amplicons were sequenced unidirectionally using Roche's 454 Titanium FLX Technology (Roche Diagnostics Nederland BV, Almere, The Netherlands). A total of 197 164 16S rRNA gene sequences were analyzed, of which 147 466 reads could be assigned to families, and 31 967 reads could be assigned to genera. We found 10 011 distinct operational taxonomic units (OTUs) in all the samples we studied.
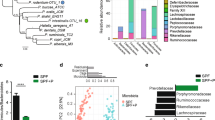
To measure how phylotype richness in the four cecal communities varied with sample size, we calculated phylotype rarefaction curves at 97% similarity levels. Richness is a measure of biodiversity and represents the number of different taxa or phylotypes (OTUs) in an ecosystem. High species richness for a given genotype generally points to a high level of redundancy in its function, which denotes the ability of the microbiome to withstand disturbances. Besides richness, also the population structure might change. To assess this, we measured the Shannon diversity index that combines the evenness of taxa distribution and the richness of each sample. The Shannon diversity is increased either by having additional unique species, or by having increased-species evenness. The average observed richness at a rarefaction depth of 5000 reads per sample for each genotype were 678 phylotypes in the microbiomes of the WT mice and 606, 682, and 650 phylotypes, respectively, in the Casp-3-, -1- and -7-KO mice. In our dataset, the four microbial communities did not differ significantly in phylotype richness (P=0.37, Figure 1a) or Shannon diversity (P=0.26, data not shown). Although we did not observe significant differences at the OTU level, we observed some trends that proved to be significantly different at the higher taxonomic genus and family levels. At these levels, our analysis revealed significant differences between the microbiome richness of the four genotypes (P=0.016 genus, 0.047 family; Supplementary Figures S1a and b). The observed genus and family richness of communities from Casp-3-KO mice was significantly less than in the WT, Casp-1 and Casp-7-KO mice. Shannon diversity analysis at the genus and family levels revealed a significant general pattern in average diversity: Casp-3-KO < WT < Casp-1 KO, Casp-7 KO (P=0.032 genus, 0.023 family, Supplementary Figures S1c and d).
Ecological community analysis of identified 16S rRNA sequences. (a) Rarefaction curves were used to estimate richness of the microbiota belonging to caspase-KO and -WT mice. The vertical axis shows the number of OTUs observed after rarefying samples to a given read depth, as indicated on the horizontal axis. (b) 3D NMDS of Bray–Curtis distance between samples. Dimensions 1 and 3 are shown. Each symbol represents one sample. The variance explained by each dimension is indicated in parentheses on the axes. Dark red, Casp-7 KO; red, Casp-1 KO; orange, Casp-3 KO; blue, WT
The difference between the OTU and the higher taxonomic levels may be explained by the higher probability of richness estimation deviations due to sequencing errors and short-read length at the OTU level.23 Therefore, we decided to focus the analyses in this study on higher-level taxonomic assignments.
Next, we compared the overall bacterial community composition using non-metric multidimensional scaling (NMDS) to explore similarities and dissimilarities between samples. The NMDS algorithm starts with a matrix of data similarities and then assigns a location to each data point in the N-dimensional space, using a non-linear least squares fit to minimize the distances between the data points. The resulting data positions can be displayed in three dimensions. Dimensional reduction of the pairwise Bray–Curtis distance between samples at the family level had a good fit with a stress of 4. NMDS dimensions 1 and 3 showed a partially overlapping clustering of the genotypes (Figure 1b), with a clear separation between Casp-3-KO samples, and Casp-1- and -7-KO samples. The first NDMS component explained 54.3% of the total variance among the individual samples. We next tested the inter-sample Bray–Curtis distances for each group of samples (insert Figure 1b). This analysis demonstrated that the WT samples form the tightest cluster, indicating similar WT sample composition, whereas the Casp-3-KO samples are more scattered. From the NMDS analysis, we conclude that the microbial communities of Casp-7 and Casp-3 KO are the least alike, and that microbiomes of WT and Casp-1 KO cluster in an intermediary position.
Distinctive gut microbial communities are associated with caspase deficiency
Although NMDS provides a good first overview of sample similarities, it does not point out specific differences between samples. Therefore, we tested whether the abundance of certain phyla, classes, families and genera were significantly different when comparing the four genotypes (Supplementary Table S2). Most of the significant differences were observed between the microbiomes of WT and Casp-7-KO mice, and when comparing the microbiomes of Casp-3-KO with Casp-1-KO or Casp-7-KO mice.
At the phylum level, the majority of the sequences were distributed among seven bacterial phyla. Firmicutes and Bacteroidetes together harbored more than 80% of the sequences, whereas Proteobacteria, Tenericutes, Deferibacteres, Actinobacteria and TM7, each accounted for 0.1–5% of sequences. Figure 2a shows the distribution of these phyla for each mouse that was used for sequencing. The dominance of the Firmicutes and Bacteroidetes phyla in our mouse gut microbiomes is in agreement with other studies.19 The relative abundance of these phyla, however, was altered in the Casp-3-, -7- and -1-KO groups compared with the WT group. Firmicutes represented a significantly lower proportion in Casp-7 and Casp-1 mice. On average, Firmicutes were 24% (P=0.008, q=0.03) and 12% (P=0.03, q=0.14) less abundant in these mice than in WT mice, respectively. By contrast, Casp-3 mice carried significantly more Firmicutes, about 8% more than WT (P=0.05, q=0.15). The pattern of Bacteroidetes abundance appeared to be in the opposite direction; Casp-7 and Casp-1-KO mice had more Bacteroidetes than WT mice (39% and 18% respectively), whereas Casp-3-deficient mice carried 12% less Bacteroidetes than WT mice. This difference was only significant for Casp-7-KO mice (P=0.006, q=0.005).
Abundance of bacterial phyla in caspase-KO and -WT mice. (a) Abundance plot of the most important phyla. Each horizontal line represents one mouse. (b) Ratio of Firmicutes/Bacteroidetes as determined by sequencing analysis; (c) Ratio of Firmicutes/Bacteroidetes as determined by qPCR. The ratio of a WT mouse was set at 1, and then the relative ratios for all genotypes were calculated. Shown are mean and S.E.M. Number of samples (n) are shown below the figures. Differences between WT and KO samples were tested by t-test. *P<0.05, **P<0.01, ***P<0.005 versus WT
The ratio of Bacteroidetes to Firmicutes might be important for a healthy intestinal bacterial community and has been described several times, for example, in the context of obesity and aging.18, 24 To validate the results obtained by sequencing (Figure 2b), we extended the initial sample set of five mice per genotype with a larger independently obtained set of samples. We confirmed our sequencing results by performing a quantitative PCR (qPCR) specific for Firmicutes and Bacteroidetes to be able to calculate the Firmicutes/Bacteroidetes (F/B) ratio of this extended sample set (Figure 2c). When the ratio of WT mice set at 1 for both techniques, the average F/B ratios for Casp-3- and -1-KO mice were 1.29 and 0.76, respectively, for sequencing, and 1.30 and 0.70, respectively, for qPCR. Although the F/B ratios in the Casp-7-KO samples were significantly lower than in WT, the difference with WT samples was less prominent in the larger data set (average ratio 0.55, n=5 versus 0.79, n=23). The pathophysiological significance of an F/B shift is not well understood. The precise composition of the species or OTUs represented by these two phyla, the detailed shifts in this composition upon treatment or disease, and/or the metabolic profile of the bacteria are certainly of importance to value this ratio.
In-depth analysis of the microbial flora at the family level allowed us to further dissect the differences between the genotypes (Figure 3a). Within the Firmicutes phylum, most sequences grouped into the Lachnospiraceae family, which is placed phylogenetically within clostridial cluster XIVa. The majority of the Bacteroidetes sequences belonged to the Porphyromonadaceae and Ruminococcaceae families. The differences in the abundance of the Bacteroidetes and Firmicutes phyla in caspase-KO and -WT mice were traced back to significant differences in the abundance of members of the Lachnospiraceae, Porphyromonadaceae and Prevotellaceae families, as well as to a large group of unclassified bacteria (Figure 3b). Other groups that differed significantly between WT and Casp-7-KO mice at the family level consisted of unclassified Firmicutes (P=0.03, q=0.15), Bacteroidetes (P=0.22, q=0.10) and Alphaproteobacteria (P=0.04, q=0.15). However, the number of sequences that represent these groups was relatively small.
Abundance of bacterial families in caspase-KO and -WT mice. (a) Abundance plots of the most abundant families. The designation ‘Other’ summarizes low-abundant families that are not explicitly shown in this graph. (b) Families with the most significantly different abundances. Shown are mean and S.E.M. Differential abundance between groups of samples was tested by t-test. *P<0.05, **P<0.01, ***P<0.005 versus WT
The Lachnospiraceae, Porphyromonadaceae and Prevotellaceae families are known for their association with intestinal inflammation. Porphyromonadaceae were significantly decreased in mice susceptible to Salmonella-induced colitis; thus, Porphyromonadaceae might be protective organisms.25 This idea is supported by the report of Wlodarska et al.,26 who suggested that a reduction of Porphyromonadaceae in the intestine contributes to the exacerbated severity of C. rodentium-induced colitis in mice that were pretreated with the antibiotic metronidazole. The significant increase of this family in Casp-1- and -7-KO mice might mean that these mice are protected in bacteria-induced colitis models, whereas the Casp-3-KO mice, which have significantly fewer Porphyromonadaceae, would be sensitized in these models. The Lachnospiraceae family was also found to be associated with the development of colitis and possibly, predisposition to colitis.25, 27 The main benefit of Lachnospiraceae family members in the human intestine is their participation in carbohydrate fermentation into butyrate (a short-chain fatty acid (SCFA)) and gas.28 SCFAs are important sources of energy for colonic epithelial cells and may enhance epithelial barrier integrity, and modulate the gastro-intestinal immune response.29 SFCAs can also induce autophagy to delay mitochondria-mediated apoptotic cell death.30 The decrease of Lachnospiraceae in Casp-7-KO mice could mean that they have less butyrate in their intestine and thus benefit less from the positive effects of this SCFA. The Prevotellaceae family contains carbohydrate- and protein-fermenting bacteria, as well as producers of acetate and hydrogen. Rho et al.31 showed that some Prevotellaceae produce sulfatases that actively degrade mucus oligosaccharides, and as a result, might disrupt the mucosal barrier in the intestine. Only very recently, Elinav et al.10 reported on a ‘colitogenic’ gut microbiome that is established by the genetic make-up of the host. The altered fecal ‘colitogenic’ microbiota of NLRP6-KO mice was characterized by an overrepresentation of Prevotellaceae.
It appears that the effect of gene deficiency on the microbiota adds another layer of regulation that affects inflammation resulting from intestinal barrier disruption. In other words, gene defects can probably predispose the organism to a certain phenotype, for example, sensitivity to inflammation, by affecting the composition of the microbiome. Our finding that Prevotellaceae are highly abundant in the Casp-1-KO mice, together with the knowledge that Casp-1 is part of the NLRP6 inflammasome, and that Casp-1-KO mice are more sensitive to DSS-colitis,7 suggest a possible link between the presence of this bacterial family and the severity of DSS-induced colitis. Therefore, the significant decrease of Prevotellaceae in the Casp-3-KO mice suggests that these mice are more resistant to DSS effects, compared with the Casp-1-KO mice.
In this paper, we describe a clear link between caspase deficiency and gut microbiota composition. Given the nature of the families that differed most in abundance between WT- and caspase-KO mice, and their reported associations with intestinal inflammation or disease, it is tempting to speculate about their role as ‘predisposing’, ‘colitogenic’ or ‘protective’ bacteria. As yet, it is very difficult to directly link changes in caspase activity with microbial modifications in the human gut, as the composition of the intestinal microflora is defined by a plethora of factors. Although microbiome studies in humans have resulted in an increased awareness regarding the importance of host–microbe interactions in health and disease, it remains challenging to determine the effect of isolated host factors on shaping the human microbial flora. Our results warrant more in-depth studies of the characteristics of the microbiome of caspase-deficient mice to associate their microbiome composition with disease phenotypes.
Materials and Methods
Animals
Casp-3- and Casp-1-KO mice have been described.22, 32 Casp-7-KO mice were generated in our laboratory (submitted for publication). KO mice and their WT littermates were raised under a 12-h light cycle in a specific pathogen-free state. All mice were born and raised in the same room of our animal facility and were given food and water ad libitum. All genotypes were backcrossed at least 10 times to a C57/BL6 background. For all experiments, mice were randomly housed together for 2 weeks to diminish possible cage effects.33 Mice were killed at the same time at the age of 8–12 weeks. Animal experiments were performed under protocols approved by the ethics committee of Ghent University.
Fecal DNA extraction
Ceca were obtained immediately after the mice were killed by cervical dislocation. The content of each intact cecum was extruded manually and frozen immediately until further use. DNA was extracted from the cecal contents using the QIAamp DNA Stool Mini Kit (Qiagen, Venlo, The Netherlands).
PCR amplification of 16S rRNA genes
16S rRNA genes were amplified using two sets of a composite forward primer and a reverse primer containing a unique 10-base multiplex identifier sequence (MID, Roche TCB Number 005-2009). The schematic primer sequences are shown in Supplementary Table S1. The forward primer of one set consisted of adaptor A (5′-CGTATCGCCTCCCTCGCGCCA-3′), a four-base linker sequence (TCAG) and a 16S rRNA primer (454B_375f 5′-CCTACGGGAGGCAGCAG-3′). The matching reverse primer consisted of adaptor B (5′-CTATGCGCCTTGCCAGCCCGC-3′), a four-base linker sequence (TCAG), and the MID sequence followed by a 16S rRNA primer (454A_926r 5′-CCGTCAATTCMTTTRAGT-3′). For the other set, adaptor A was attached to the reverse primer, and adaptor B was attached to the forward primer (Supplementary Table S1). The 16S rRNA primers amplify the V3-5 region and were previously described.34 Amplification with both sets of primers allowed us to couple the amplicons to both ‘A’ and ‘B’ beads to use a complete Roche amplification kit for unidirectional sequencing. Each DNA sample was amplified with both primer sets along with a no-template control, and the quality of the amplicons was checked on a 2% agarose gel. The amplicons were purified using the QIAquick PCR Purification Kit (Qiagen, Venlo, The Netherlands) and pooled in equimolar ratios to obtain a single sample containing all amplicons for each primer set. The samples were run on an Agilent Bioanalyzer and displayed a single peak of about 660 bp. The samples were pyrosequenced using a Roche 454 Life Sciences Genome Sequencer FLX machine at the VIB Department of Molecular Genetics, University of Antwerp. The GS FLX Titanium SV emPCR kit (Lib-A) was used for titrations, and the GS FLX Titanium MV emPCR kit (Lib-A) was used for amplification of DNA libraries. For pyrosequencing, the GS FLX Titanium Sequencing kit was used (Roche).
Sequence analysis
Sequences were analyzed with the QIIME pipeline, version 1.1.35 After multiplexed sequencing of the 16S PCR products, sequences were assigned to samples on the basis of their MID tag, allowing for one base error. Only 454 reads with a length >200 bp and <1000 bp, an average q-value above 25, fewer than two ambiguous bases, and fewer than two primer mismatches were retained for further analysis. To remove sequencing errors, chimeric reads were identified and removed using ChimeraSlayer34 with default settings. Chimera-cleaned reads were denoised using Denoiser 0.84 (freeware; http://www.microbio.me/denoiser/), and OTUs were subsequently clustered from denoised reads at a 97%-identity threshold using Uclust with QIIME default settings.36, 37 We retained 197 164 high-quality reads for further analysis, with an average of 9858 reads per sample. For each OTU, the most abundant longest sequence served as a representative read. Taxonomic assignments of reads were done with RDP classifier38 with an 80% confidence limit, and were considered unassigned if below this limit. Abundance matrices of phylogenetic abundance per sample were calculated at different taxonomic levels from OTU and respective taxonomic classifications features.
Statistical analysis
To make comparisons, sample-wise feature abundances (i.e. the abundance of different phyla, classes, families, genera and OTUs in different samples) derived from the QIIME pipeline were statistically analyzed on normalized feature-abundance matrices using R 2.11.0 (freeware; http://www.r-project.org/). For normalization, all features within a sample were divided by the respective sample's sum over all features. To get an approximate representation of the feature-read depth, all proportions were multiplied by the average-read depth over all samples. As a result, samples are normalized for differences in sequencing depth, whereas providing an approximation to the number of reads assigned to each taxonomic group. Feature-abundance matrices were transformed by adding 1 to each feature and calculating log10, avoiding negative-infinite values for absent features. To investigate overall similarities and differences between samples, pairwise Bray–Curtis distances between samples were visualized using NMDS ordination, and calculated using Vegan version 1.17-9 (freeware 2011; http://vegan.r-forge.r-project.org/). The significance of differential abundance between groups of samples was calculated on the transformed feature abundances using a t-test (P-value) that was corrected for multiple testing by calculating the Benjamini–Hochberg false discovery rate (q-value). Taxa with less than 10 reads present over all normalized samples were filtered out before univariate testing.
To calculate the observed phylogenetic richness (i.e. the number of different species, genera, families) of our data set, samples were rarefied to the size of the smallest sample to allow comparison between samples of different read depth. Subsequently, the number of OTUs in each sample was calculated. The rarefaction and observed richness calculation was repeated five times, and the average of these repetitions was used. Likewise, Shannon diversity, which also takes the population structure into account, was calculated on the rarefied matrices and reflects the average diversity over five repetitions. The Kruskal–Wallis rank sum test was used to test for differences in the observed richness and Shannon diversity between groups of samples.
Real-time PCR analysis
qPCR amplification and detection were performed using the Roche Light Cycler 480 machine with the Roche LightCycler 480 SYBR Green I Master kit. The primers are listed in Supplementary Table S1. Each PCR reaction included 300 nM of forward and reverse primer, 9 ng of DNA, and SYBR Green Master Mix. Samples without template served as negative controls. Samples were incubated at 95 °C for 5 min, and then amplified through 45 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 1 s. The amount of Firmicutes and Bacteroidetes 16S rRNA in each sample was normalized to the total amount of 16S rRNA amplified with universal primers.39, 40
Abbreviations
- Casp-1:
-
caspase-1
- Casp-3:
-
caspase-3
- Casp-7:
-
caspase-7
- WT:
-
wild-type
- KO:
-
knockout
- IL-1:
-
interleukin 1
- IL-18:
-
interleukin 18
- IL-33:
-
interleukin 33
- DSS:
-
dextran sulfate sodium
- MID:
-
multiplex identifier sequence
- OTU:
-
operational taxonomic unit
- NMDS:
-
non-metric multidimensional scaling
- qPCR:
-
quantitative PCR
- SCFA:
-
short-chain fatty acid
References
Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P . Caspases in cell survival, proliferation and differentiation. Cell Death Differ 2007; 14: 44–55.
Demon D, Van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J et al. Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol Cell Proteomics 2009; 8: 2700–2714.
Luthi AU, Cullen SP, McNeela EA, Duriez PJ, Afonina IS, Sheridan C et al. Suppression of interleukin-33 bioactivity through proteolysis by apoptotic caspases. Immunity 2009; 31: 84–98.
Kobori A, Yagi Y, Imaeda H, Ban H, Bamba S, Tsujikawa T et al. Interleukin-33 expression is specifically enhanced in inflamed mucosa of ulcerative colitis. J Gastroenterol 2010; 45: 999–1007.
Mitchell JA, Paul-Clark MJ, Clarke GW, McMaster SK, Cartwright N . Critical role of toll-like receptors and nucleotide oligomerisation domain in the regulation of health and disease. J Endocrinol 2007; 193: 323–330.
Netea MG, Simon A, van de Veerdonk F, Kullberg BJ, Van der Meer JW, Joosten LA . IL-1beta processing in host defense: beyond the inflammasomes. PLoS Pathog 2010; 6: e1000661.
Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 2010; 32: 367–378.
Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD . The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010; 32: 379–391.
Siegmund B, Lehr HA, Fantuzzi G, Dinarello CA . IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. Proc Natl Acad Sci USA 2001; 98: 13249–13254.
Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011; 145: 745–757.
Rolli J, Loukili N, Levrand S, Rosenblatt-Velin N, Rignault-Clerc S, Waeber B et al. Bacterial flagellin elicits widespread innate immune defense mechanisms, apoptotic signaling, and a sepsis-like systemic inflammatory response in mice. Crit Care 2010; 14: R160.
Akhter A, Gavrilin MA, Frantz L, Washington S, Ditty C, Limoli D et al. Caspase-7 activation by the Nlrc4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS Pathog 2009; 5: e1000361.
Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX et al. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol 2011; 4: 279–287.
Garrison SP, Thornton JA, Hacker H, Webby R, Rehg JE, Parganas E et al. The p53-target gene puma drives neutrophil-mediated protection against lethal bacterial sepsis. PLoS Pathog 2010; 6: e1001240.
Rose FR, Bailey K, Keyte JW, Chan WC, Greenwood D, Mahida YR . Potential role of epithelial cell-derived histone H1 proteins in innate antimicrobial defense in the human gastrointestinal tract. Infect Immun 1998; 66: 3255–3263.
Scharl M, Rogler G . Microbial sensing by the intestinal epithelium in the pathogenesis of inflammatory bowel disease. Int J Inflam 2010; 2010: 671258.
Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al. Enterotypes of the human gut microbiome. Nature 2011; 473: 174–180.
Ley RE, Turnbaugh PJ, Klein S, Gordon JI . Microbial ecology: human gut microbes associated with obesity. Nature 2006; 444: 1022–1023.
Spor A, Koren O, Ley R . Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 2011; 9: 279–290.
Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI . Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One 2008; 3: e3064.
Lakhani SA, Masud A, Kuida K, Porter Jr GA, Booth CJ, Mehal WZ et al. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 2006; 311: 847–851.
Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell 1995; 80: 401–411.
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P . Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 2010; 12: 118–123.
Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 2009; 9: 123.
Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA et al. The intestinal microbiota plays a role in salmonella-induced colitis independent of pathogen colonization. PLoS One 2011; 6: e20338.
Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun 2011; 79: 1536–1545.
Lepage P, Hasler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011; 141: 227–236.
Cotta M, Forster R . The family lachnospiraceae, including the genera Butyrivibrio, Lachnospira and Roseburia. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds). The Prokaryotes. Springer: New York, 2006, pp. 1002–1021.
Meijer K, de Vos P, Priebe MG . Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Curr Opin Clin Nutr Metab Care 2010; 13: 715–721.
Tang Y, Chen Y, Jiang H, Nie D . Short-chain fatty acids induced autophagy serves as an adaptive strategy for retarding mitochondria-mediated apoptotic cell death. Cell Death Differ 2011; 18: 602–618.
Rho JH, Wright DP, Christie DL, Clinch K, Furneaux RH, Roberton AM . A novel mechanism for desulfation of mucin: identification and cloning of a mucin-desulfating glycosidase (sulfoglycosidase) from Prevotella strain RS2. J Bacteriol 2005; 187: 1543–1551.
Leonard JR, Klocke BJ, D’Sa C, Flavell RA, Roth KA . Strain-dependent neurodevelopmental abnormalities in caspase-3-deficient mice. J Neuropathol Exp Neurol 2002; 61: 673–677.
Fushuku S, Fukuda K . Inhomogeneity of fecal flora in separately reared laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE). Exp Anim 2008; 57: 95–99.
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 2011; 21: 494–504.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7: 335–336.
Reeder J, Knight R . Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 2010; 7: 668–669.
Edgar RC . Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26: 2460–2461.
Wang Q, Garrity GM, Tiedje JM, Cole JR . Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73: 5261–5267.
Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K . Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol 2008; 47: 367–373.
Fierer N, Jackson JA, Vilgalys R, Jackson RB . Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 2005; 71: 4117–4120.
Acknowledgements
Research in the Vandenabeele group has been supported by European grants (FP6 ApopTrain, MRTN-CT-035624; FP7 EC RTD Integrated Project, Apo-Sys, FP7-200767; Euregional PACT II), Belgian grants (Interuniversity Attraction Poles, IAP 6/18), Flemish grants (Fonds Wetenschappelijk Onderzoek ‘FWO’ Vlaanderen, G0226.09, 1.5.169.08, G.0875.11 and G.0973.11), Ghent University grants (MRP, GROUP-ID), and grants from the Flanders Institute for Biotechnology (VIB). PV holds a Methusalem grant (BOF09/01M00709) from the Flemish Government. JR and FH are supported by the Institute for the encouragement of Scientific Research and Innovation of Brussels (ISRIB) and the Odysseus program of the FWO. We thank Dr. Amin Bredan (DMBR-VIB, Ghent) for editing the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Piacentini
Supplementary Information accompanies the paper on Cell Death and Disease website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Brinkman, B., Hildebrand, F., Kubica, M. et al. Caspase deficiency alters the murine gut microbiome. Cell Death Dis 2, e220 (2011). https://doi.org/10.1038/cddis.2011.101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2011.101
Keywords
This article is cited by
-
Bacterial communities associated with mushrooms in the Qinghai-Tibet Plateau are shaped by soil parameters
International Microbiology (2022)
-
Development of a Pumpless Microfluidic System to Study the Interaction between Gut Microbes and Intestinal Epithelial Cells
Biotechnology and Bioprocess Engineering (2022)
-
Dock2 affects the host susceptibility to Citrobacter rodentium infection through regulating gut microbiota
Gut Pathogens (2021)
-
CD36 deficiency affects depressive-like behaviors possibly by modifying gut microbiota and the inflammasome pathway in mice
Translational Psychiatry (2021)
-
Residual feed intake divergence during the preweaning period is associated with unique hindgut microbiome and metabolome profiles in neonatal Holstein heifer calves
Journal of Animal Science and Biotechnology (2020)