Abstract
We have recently found that primary rat embryonic fibroblasts (REFs) could be immortalized by overexpression of the human mitochondrial ribosomal protein MRPS18-2 (S18-2). A derived cell line, designated 18IM, expressed the embryonic stem cell markers SSEA-1 and Sox2. Upon inoculation into severe combined immunodeficiency mice, 18IM cells differentiated to express pan-keratin. They were not tumorigenic. Here we report the gene profiling of 18IM, compared with REF cells. Pathways involved in oxidative phosphorylation, ubiquinone (Coenzyme Q 10) biosynthesis, fatty acid elongation in mitochondria, PI3K/AKT signaling, a characteristic of rapidly proliferating cells, were upregulated in 18IM. Genes involved in the transcription/translation machinery and redox reactions, like elongation factors, ATP synthases, NADH dehydrogenases, mitogen activated kinases were upregulated as well. 18IM cells produced more pyruvate, indicating enhanced ATP synthesis. The expression of Oct4, Sox2, and Nanog that can contribute to the experimental induction of pluripotency in primary fibroblasts was also elevated, in contrast to Klf4 and C-myc that were downregulated. Subsequently, three new immortalized cell lines were produced by S18-2 overexpression in order to check the representativeness of 18IM. All of them showed anchorage-independent growth pattern. Two of three clones lost vimentin and smooth muscle actin, and expressed Sox2 and Oct4. We suggest that S18-2 is involved in the developmental regulation.
Similar content being viewed by others
Main
Mouse embryonic fibroblasts have been converted into pluripotent stem cells (iPSC) in vitro, by expressing four genes, SOX2 (NM_003106), OCT4 (NM_002701), KLF4 (NM_004235), and C-MYC (NM_002467)1, 2, 3 or, alternatively, N-MYC (NM_005378).4 Rat fibroblasts could also be turned into iPSC by the same four genes.5 Human primary fibroblasts could be converted into stem cells by the same or a slightly different set of genes, such as OCT4, SOX2, NANOG (XM_002344645), and LIN28 (NM_024674).6 Also, OCT4 and SOX2 could induce pluripotency in human fibroblasts by themselves, in conjunction with valproic acid, a histone deacetylase inhibitor.7 SOX2 and OCT4 could generate iPSCs from cord blood cells without valproic acid.8, 9
We have accidentally discovered that overexpression of the human mitochondrial ribosomal protein MRPS18-2 (S18-2, NP_054765) immortalized primary rat embryonic fibroblasts (REFs).10 The derived cell line, designated 18IM, lost its sensitivity to contact inhibition and acquired the ability for anchorage-independent growth in soft agar with a cloning efficiency of 94%. It expressed the embryonic stem cell markers SSEA-1 and Sox2 that were not detected in the original REFs, nor in C-MYC- and HA-RAS-transformed REFs. Moreover, the 18IM cells lost the expression of mesodermal markers like vimentin and smooth muscle actin (SMA). In contrast, they turned on the ectoderm- and endoderm-specific pan-keratin, ectoderm-specific beta-III-tubulin, and mesoderm-specific MHC class II markers in confluent culture. Part of the cells differentiated into Oil red O stainable fat cells.
Upon subcutaneous inoculation into severe combined immunodeficiency (SCID) mice, 18IM cells differentiated to express pan-keratin. Unlike C-MYC and HA-RAS-transformed fibroblasts, they were not tumorigenic.10
Here we report the gene expression pattern of 18IM cells and three new S18-2-immortalized cell lines, in comparison with control REFs.
Results
Stem cell marker expression in 18IM cells
The gene expression profile of 18IM cells was compared with REFs by microarray techniques. Genes that showed a twofold and higher difference with FDR-corrected P-value <0.05 were selected for further studies.
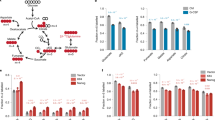
Supplementary Table S1 shows the differential gene expression. More than 4000 differently expressed genes are shown in an overview (Figure 1).
Ingenuity Pathway Analysis (IPA, Ingenuity Systems Inc., Redwood City, CA, USA) was performed for Gene ontology studies on differential expression set (4209 genes). Several canonical pathways, including ‘human embryonic stem cell pluripotency’, the ‘NANOG pathway’, and others were activated in 18IM cells (Table 1).
At least 19 cellular pathways were altered in the S18-2-immortalized cells (Table 1). Upregulated genes include COX and NDUF family genes, involved in proliferation, oxidative phosphorylation, cellular respiration, and other redox reactions, suggesting that 18IM cells are metabolically more active. Their increased pyruvate release suggested an elevated level of ATP production (Supplementary Figure S1), in accordance with higher proliferation rate, as reported earlier.10
Genes involved in the WNT, BMP, and other self-renewal pathways, like β-catenin (Ctnn-β), Bmpr3 (BMP receptor), Smad1/5/8, Stat3, Spp1, Sox11, and Sh3glb1 were upregulated. In contrast, genes involved in mesenchymal differentiation, such as Runx2, Hoxc8, Creb, and Tlx2, or endoderm development, as Foxd3, were downregulated. Essential factors in the TGF-β and BMP pathways, like TGFFB3, BMP1, BMP7, SMAD1, and CREB1 were also upregulated with pathway expectation (−log (P-value)) of 1.64 and 1.19, respectively. Many genes involved in the regulation of stem cell development and proliferation were differently expressed in 18IM and REF cells. Genes involved in the proliferation of embryonic cells (30 genes), proliferation of neural stem cells (9 genes), and survival of embryonic cells (11 genes) were also differentially expressed.
Quantitative PCR validation of microarray data
Fifteen genes that have an important role in stem cell phenotype maintenance were chosen to validate the microarray data by quantitative PCR (Q-PCR). The elongation factors Eef1A2 (NM_012660) and Tcea3 (NM_001015008) were strongly upregulated in 18IM cells, in agreement with the microarray data (Figure 2). The expression of Eef1A2 increased 10 000-fold (8933 according to microarray) and of Tcea3 increased by 393-fold (500 by microarray), suggesting enhanced RNA synthesis. Gata3 (NM_133293) that may be involved in histone acetylation11 was overexpressed (Figure 2, middle panel) while Gata4 (NM_144730) and Gata6 (NM_019185), which are usually expressed in differentiated tissues, were downregulated (Figure 2, lowest panel). Genes influencing cell proliferation like Tiam1 (NM_001100558), Epas1 (NM_023090), and Jmjd4 (NM_001105784) were expressed at a higher level in 18IM cells. Similar changes were recorded by Q-PCR and microarray for Tiam1 (566/106) and Jmjd4 (98/145).
18IM cells also showed a high expression of the testis-specific Tssk1 gene (NM_001011900).12 EST data showed that this gene is expressed in the brain and in the germ cell tumors. The placenta-specific Cdx2 gene (NM_023963) was 100 times more highly expressed in 18IM, compared with REF cells. Cdx2 expression was also reported in gastrointestinal and colorectal tumors, gliomas, and embryonic tissue. Genes that participated in the experimental induction of stem cell markers in normal fibroblasts, such as Oct4, Nanog, and Sox2 were also more highly expressed in 18IM cells. However, C-myc and Klf4 were downregulated in the immortalized cells.
There was no significant change in the methylation status of the promoter region of Oct-4, Sox2, Nanog, Lin28, and C-myc in 18IM cells compared with REFs (see Supplementary Table S3), nor was there any difference in total genome methylation (Supplementary Figure S2).
Protein expression
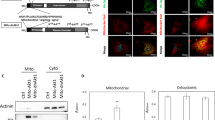
Protein expression was assayed by western blotting and immunostaining. The immortalized 18IM cells have lost vimentin and SMA expression, concomitantly with the rise of Sox2, HIF1A, p53, RB, and p14ARF protein levels, confirming our previous data (Figure 3). Despite lower levels of C-myc mRNA, C-myc protein was expressed at the higher level in 18IM cells, compared with REFs, probably, due to protein stabilization.
It is noteworthy that the embryonic stem cell marker SSEA-1 was expressed on 30% of 18IM cells that have grown in culture for >2 years (Supplementary Figure S2A). They were non tumorigenic in SCID mice, after the inoculation of 0.5–2 × 106 cells.
Experiments on newly generated S18-2-immortalized REF lines
Three new S18-2-immortalized cell lines were generated by transfecting REFs with GFP-S18-2 and pBabe-S18-2 plasmids, followed by selection on G418. They were followed in culture for 12 months. Clones 10 and 12 that expressed GFP-S18-2 at a high level grew similarly to 18IM cells, forming foci with a dense center, whereas clone 6 that expressed a much lower level of S18-2 protein from a pBabe construct did not form compact foci, but grew as a monolayer, showing a high degree of cellular heterogeneity. Clones 10 and 12 but not clone 6 showed anchorage-independent growth (Figure 4). Cells (0.5 × 106) were inoculated subcutaneously in SCID mice. After 2 months no tumor formation was detected.
Growth pattern of clones produced from REFs that expressed S18-2 protein at different levels. Growth of clone 10 (a, GFP-S18-2) and clone 12 (b and c, GFP-S18-2) in bacterial Petri dish. Notice embryoid-like body formation and proliferation of adherent cells. Growth of clone 6 (d–f), expressing S18-2 from pBabe construct, in the ordinary flask. Notice the heterogeneity of the cells, produced from one clone: smaller in the middle and larger at the periphery
The protein expression pattern of the three new clones was compared with 18IM and REF cells by western blotting. Clone 6 showed no loss of vimentin and SMA, in contrast to 18IM and clones 10 and 12 that expressed S18-2 at a high level (Figure 5). Clones 10 and 12 expressed high levels of Oct4 and Sox2, and about 5% of them expressed SSEA-1. Only the 18IM cells (that were cultured for 12 months) expressed E-cadherin.
Study of protein expression in the three recent clones, compared with 18IM cells and REFs, by western blotting. Notice that OCT4 and C-myc were expressed at a higher level in clones 10, 12, and in 18IM cells. Clone 6 that expressed S18-2 protein at a low level showed a decreased expression of SMA in comparison with parental REFs. 18IM cells expressed E-cadherin that was absent in REFs, whereas the 3 clones did not
Discussion
The MRPS18-2 (MRSPS18B, S18-2) protein is encoded by a gene on human chromosome 6p21. 3, adjacent to the MHC class II gene cluster. It was first cloned from CD34+ hematopoietic stem/progenitor cells by Zhang et al.13 Similarly to the other two members of the S18 family, S18-2 is localized on the surface of the small subunit (28S) of the mammalian mitochondrial ribosome.14, 15 We have previously found that S18-2 is involved in the regulation of the RB pathway. It binds to both hypo- and hyper-phosphorylated retinoblastoma protein (RB) but not to other pocket proteins, such as p107 and p130.16 In EBV-transformed lymphoblastoid cells a great portion of the normally cytoplasmic S18-2 is targeted to the nucleus by one of the virally encoded transforming proteins, EBNA-6.17 Its binding to RB displaces E2F1, leading to an increase of free E2F1. This may presumably lift the RB-dependent block of S-phase entry; a regular corollary of EBV-induced B-cell transformation. It is noteworthy in this context that the S18-2 protein is mainly localized in the nucleus in immortalized REFs.10
We now report that numerous pathways, characteristic for rapidly proliferating cells, are upregulated in 18IM. They include genes involved in the transcription/translation machinery, such as the elongation factors Eef1A2 and Tcea3, and enzymes that participate in metabolic and redox reactions, like the cytochrome C oxidases Cox4, Cox6A2, Cox4I1-2, Cox8, and the NADH dehydrogenases NdufA3, 6, 11, 13, NdufB2 and 8 (Table 1). SOD2 (superoxide dismutase), GPX4, and GPX7 (glutathione peroxidases) genes were also upregulated in 18IM cells. Moreover, ATP synthases (ATP5-E, F1, H, I, J2, and ATP50, ATP6-V0A1, and ATP6-V1E1), usually located in the inner membrane of the mitochondria, were also elevated in 18IM cells.
They also showed increased release of pyruvate (Supplementary Figure S1) that may be associated with enhanced ATP production.
The expression of Oct4, Sox2, and Nanog that have been used for the experimental induction of PSC differentiation in primary fibroblasts was elevated in 18IM (Figure 2). Other genes in the same experimental category,5 such as Klf4 and C-myc, were downregulated in 18IM cells. However, C-myc protein was expressed at the higher level in immortalized cells. It is noteworthy that 18IM cells was unable to produce tumors in SCID mice, they transdifferentiated.10 We may speculate that C-myc protein was stabilized in immortalized cells but its transactivating ability was inhibited.
As a step towards exploring the developmental role of the S18-2 protein, we inhibited its expression in the Danio rerio (Zebrafish) model. Introduction of a S18-2-specific morpholino into the fish eggs led to the block of S18-2 translation. This resulted in embryonic lethality (24–72 h). Forty-eight hours after the introduction of the morpholino into 308 eggs left no living fish, in contrast to >88% of normally looking fish in the control experiments. Simultaneous injection of morpholino and in-vitro-transcribed S18-2 mRNA permitted the development of normal fish. Further studies are in progress to explore the developmental role of S18-2 in development in zebrafish, xenopus, and mouse models.
Materials and Methods
Cell cultures
Primary REFs and 18IM cells were cultured at ordinary conditions at 37°C, using Iscove's Modified Dulbecco's Medium (IMDM) medium supplemented by 10% of fetal bovine serum and appropriate antibiotics. 18IM were cultured without feeder layer in IMDM that contained 0.5 mg/ml of G418. For microarray analysis 18IM cells were used that were propagated for more than 1 year. Hoechst staining was used to monitor the absence of mycoplasma.
Microarray analysis
Total RNA was extracted from samples using the RNeasy Mini kit (Qiagen Inc., Hilden, Germany), according to the manufacturer′s recommendations. For each sample, 200 ng total RNA was reverse transcribed, linear amplified, and labeled with Cy3 using the one color Low RNA Input Linear Amplification Kit PLUS (Agilent Technologies, Santa Clara, CA, USA), according to the manufacturer's instructions and purified using the RNeasy Mini kit (Agilent Technologies). After labeling, samples were measured on a Nanodrop microarray module for labeling efficiency and quantification. Samples were then hybridized on Agilent 4 × 44K whole rat genome GE arrays (Agilent Design #014850) at 65°C for 17 h. After washing with GE washing buffer, the slide was scanned with Agilent Microarray Scanner G2565BA (Agilent Technologies) at 5-μm resolution. Feature extraction software (Version 9.5.3.1) was used to convert the image into gene expression data. Data were normalized by the quantile method, using Partek. Only genes with a false discovery rate ≤0.05 were selected as significant with fold change cutoff of 2. Molecular functions of genes were classified according to Gene Ontology function using Ingenuity Pathway Analysis (IPA).
Methylation of gene promoter region in genomic DNA by direct sequencing after sodium bisulfite treatment is described in Supplement (Materials and Methods chapter).
Real-time PCR
Total RNA was isolated from REF and 18IM cells, using RNeasy Mini kit (Qiagen Inc.). Approximately 1 μg of total RNA was used for cDNA synthesis, using First Strand Synthesis Kit (Sigma-Aldrich, St. Louis, MO, USA) according to manufacturer's protocol. cDNA obtained was used for Q-PCR studies. Primer concentration was adjusted to a final concentration of 3 μM. Total reaction volume for all real-time PCR experiments was 20 μl. Real-time PCR was performed using SYBR GREEN Master mix on 7900 machine (Applied Biosystems, Foster City, CA, USA). The following primers for rat genes were used:
Cdx2 (Caudal type homeobox 2) For 5′-CGTCCTGCGCCTCCGGATTG-3′, Rev 5′-AGGGACGGCTGAGCTGGCTT-3′; C-myc For 5′-TCTCCGTCCTATGTTGCGGTC-3′, Rev 5′-TCCAAGTAGCTCGGTCATCATCT-3′; Eef1A2 (eukaryotic translation elongation factor 1 alpha 2) For 5′-CAAGGGCTGGAAGGTAGAGCG-3′, Rev 5′-CCCAATGCCACCAATCTTGT-3′; Epas1 (endothelial PAS domain protein 1 or Hif2A) For 5′-CCCAAGAAGGTGACATGATCT-3′, Rev 5′-CGCAGGGATGAGTGAAGTCAAA-3′; Gata-3 (GATA binding protein 3) For 5′-CGAGGCCCAAGGCACGATCC-3′, Rev 5′-GCTCTCCTCGCTGCTGACAGC-3′; Gata-4 For 5′-GGAGCTGGCCAGGACTGTCG-3′, Rev 5′-TGCGCAGGCCTTCGGATCAC-3′; Gata-6 For 5′-CGGCCTGCCTTACCTGCAGG-3′, Rev 5′-GGCGAGTAGGGGAAGCGTGC-3′; Jmjd4 (jumanji domain containing 4), For 5′-AGCCTGGCGAGATGGTGTTTG-3′, Rev 5′-GCCATTGACCCAGTTGTGGT-3′; Klf4 For 5′-TTCTCCACGTTCGCGTCCGG-3′, Rev 5′-TCTCGCCAACGGTTAGTCGGGG-3′; Nanog For 5′-TTGGAACGCTGCTCCGCTCC-3′, Rev 5′-CGCCTGGCTTTCCCTAGTGGC-3′; Oct4 For 5′-GGAGGGATGGCATACTGTGGACCT-3′, Rev 5′-TCCTGGGACTCCTCGGGACTAGG-3′; Sox2 For 5′-ACTAATCACAACAATCGCGGCGGC-3′, Rev 5′-GACGGGCGAAGTGCAATTGGGA-3′; Tcea3 (transcription elongation factor A (SII), 3) For 5′-GCTGAACAGTTGCCAGATGTC-3′, Rev 5′-GCAGCCGCTTCCAGTTTTTAAT-3′; Tiam1 (T-cell lymphoma invasion and metastasis 1) For 5′-TGCTGACATACGCCCCAACT-3′; Rev 5′-CAACAGTGGTTCTCAGCCCAA-3′; Tssk1 (testis-specific serine kinase) For 5′-CCACCGCTCTATTGTCAAGAC-3′, Rev 5′-GCTCCTCTGGTTTTGATGAACT-3′.
As an internal control for standardization, we have assayed the expression of a gene encoding TATA-binding protein (Tbp, NM_003194). The following primers were used: For 5′-TTTCTTGCCAGTCTGGAC-3′, Rev 5′-CACGAACCACGGCACTGATT-3′.
The PCR cycling conditions were the following: 10 min at 95°C, 40 cycles of 10 s at 95°C and 1 min at 60°C. Applied Biosystems 7900 system software was used for analysis. Ct values were determined for the internal control (TBP) and the test genes at the same threshold level in the exponential phase of the PCR curves. Relative quantification (comparative Ct (ddCt) method) was used to compare the expression level of the test genes with the internal control. Dissociation curve analysis was performed after every run to check the specificity of the reaction. In all, 3–5 reactions (each in triplicates) were run for each gene and a standard deviation was calculated.
Western blotting
Whole-cell lysates were prepared using NP40 lysis buffer (1% NP40, 150 mM NaCl, 50 mM Tris, pH=8) with a protease inhibitor cocktail (Sigma-Aldrich). Lysates were cleared by centrifugation.
Proteins were separated by sodium SDS-polyacrylamide gel electrophoresis. After transfer the membranes were probed with mouse antibodies against actin (Sigma-Aldrich), SMA, vimentin, and p53 (DAKO, Glostrup, Denmark), C-MYC (ABCAM Ltd, Cambridge, UK), RB (BD Pharmingen, San Diego, CA, USA), and rabbit antibodies against OCT4 (Novus Biologicals, Littleton, CO, USA), SOX2 (ABCAM Ltd), HIF1A, and E-cadherin (Cell Signaling, Denver, MA, USA). The rabbit polyclonal serum against p14ARF was the kind gift of Klas Wiman (CCK, KI, Stockholm, Sweden). Secondary antibodies (anti-rabbit and anti-mouse IgG Horseradish peroxidase-conjugated) were purchased from GE Healthcare Bio-Sciences AB (Uppsala, Sweden).
Cell culture, immunostaining, imaging, and biochemical assays are described in Supplement (Materials and Methods chapter).
Abbreviations
- REF:
-
rat embryonic fibroblasts
- S18-2–MRPS18-2:
-
mitochondrial ribosomal protein S18-2
- SCID:
-
severe combined immunodeficiency
- 18IM–REFs:
-
immortalized by overexpression of MRPS18-2 (S18-2) protein
- iPSC:
-
induced pluripotent stem cells
- IMDM:
-
Iscove's Modified Dulbecco's Medium
- Q-PCR:
-
quantitative PCR
References
Takahashi K, Yamanaka S . Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676.
Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007; 448: 318–324.
Meissner A, Wernig M, Jaenisch R . Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotech 2007; 25: 1177–1181.
Blelloch R, Venere M, Yen J, Ramalho-Santos M . Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell 2007; 1: 245–247.
Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y et al. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell 2009; 4: 11–15.
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318: 1917–1920.
Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotech 2008; 26: 1269–1275.
Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodriguez-Piza I, Vassena R et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell 2009; 5: 353–357.
Giorgetti A, Montserrat N, Rodriguez-Piza I, Azqueta C, Veiga A, Izpisua Belmonte JC . Generation of induced pluripotent stem cells from human cord blood cells with only two factors: Oct4 and Sox2. Nat Prot 2010; 5: 811–820.
Kashuba E, Pavan Yenamandra S, Deoram Darekar S, Yurchenko M, Kashuba V, Klein G et al. MRPS18-2 protein immortalizes primary rat embryonic fibroblasts and endows them with stem cell-like properties. Proc Nat Acad Sci 2009; 106: 19866–19871.
Chen GY, Osada H, Santamaria-Babi LF, Kannagi R . Interaction of GATA-3/T-bet transcription factors regulates expression of sialyl Lewis X homing receptors on Th1/Th2 lymphocytes. Proc Nat Acad Sci 2006; 103: 16894–16899.
Hao Z, Jha KN, Kim YH, Vemuganti S, Westbrook VA, Chertihin O et al. Expression analysis of the human testis-specific serine/threonine kinase (TSSK) homologues. A TSSK member is present in the equatorial segment of human sperm. Mol Hum Reprod 2004; 10: 433–444.
Zhang QH, Ye M, Wu XY, Ren SX, Zhao M, Zhao CJ et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res 2000; 10: 1546–1560.
Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A et al. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J Biol Chem 2001; 276: 33181–33195.
Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL . The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem 2001; 276: 19363–19374.
Snopok B, Yurchenko M, Szekely L, Klein G, Kashuba E . SPR-based immunocapture approach to creating an interfacial sensing architecture: mapping of the MRS18-2 binding site on retinoblastoma protein. Anal Bioanal Chem 2006; 386: 2063–2073.
Kashuba E, Yurchenko M, Yenamandra SP, Snopok B, Isaguliants M, Szekely L et al. EBV-encoded EBNA-6 binds and targets MRS18-2 to the nucleus, resulting in the disruption of pRb-E2F1 complexes. Proc Nat Acad Sci 2008; 105: 5489–5494.
Acknowledgements
Our work was supported by the Swedish Cancer Society, by matching grants from the Concern Foundation (Los Angeles) and the Cancer Research Institute (New York), by the Swedish Institute, and by the Swedish Foundation for Strategic Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by RA Knight
Supplementary Information accompanies the paper on Cell Death and Disease website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Yenamandra, S., Darekar, S., Kashuba, V. et al. Stem cell gene expression in MRPS18-2-immortalized rat embryonic fibroblasts. Cell Death Dis 3, e357 (2012). https://doi.org/10.1038/cddis.2011.138
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2011.138
Keywords
This article is cited by
-
The MRPS18-2 protein levels correlate with prostate tumor progression and it induces CXCR4-dependent migration of cancer cells
Scientific Reports (2018)
-
Upregulation of Mrps18a in breast cancer identified by selecting phage antibody libraries on breast tissue sections
BMC Cancer (2017)
-
Immortalized tumor derived rat fibroblasts as feeder cells facilitate the cultivation of male embryonic stem cells from the rat strain WKY/Ztm
SpringerPlus (2014)
-
Loss of connective tissue growth factor as an unfavorable prognosis factor activates miR-18b by PI3K/AKT/C-Jun and C-Myc and promotes cell growth in nasopharyngeal carcinoma
Cell Death & Disease (2013)