Abstract
Autophagy is a cellular catabolic process by which long-lived proteins and damaged organelles are degradated by lysosomes. Activation of autophagy is an important survival mechanism that protects cancer cells from various stresses, including anticancer agents. Recent studies indicate that pyrvinium pamoate, an FDA-approved antihelminthic drug, exhibits wide-ranging anticancer activity. Here we demonstrate that pyrvinium inhibits autophagy both in vitro and in vivo. We further demonstrate that the inhibition of autophagy is mammalian target of rapamycin independent but depends on the transcriptional inhibition of autophagy genes. Moreover, the combination of pyrvinium with autophagy stimuli improves its toxicity against cancer cells, and pretreatment of cells with 3-MA or siBeclin1 partially protects cells from pyrvinium-induced cell death under glucose starvation, suggesting that targeted autophagy addiction is involved in pyrvinium-mediated cytotoxicity. Finally, in vivo studies show that the combination therapy of pyrvinium with the anticancer and autophagy stimulus agent, 2-deoxy-D-glucose (2-DG), is significantly more effective in inhibiting tumor growth than pyrvinium or 2-DG alone. This study supports a novel cancer therapeutic strategy based on targeting autophagy addiction and implicates using pyrvinium as an autophagy inhibitor in combination with chemotherapeutic agents to improve their therapeutic efficacy.
Similar content being viewed by others
Main
Macroautophagy (herein referred to as autophagy) is an evolutionarily conserved catabolic process that mediates degradation of intracellular components in lysosomes.1 Autophagy is controlled by the cellular transduction pathways that interpret the status of cellular energy and nutrients, which allows a tightly regulated autophagic response to environmental perturbations. AMP-activated protein kinase (AMPK) triggers autophagy, whereas autophagy is inhibited by the mammalian target of rapamycin (mTOR).2 Recent studies have suggested an important role of Vps34 complex in manipulating both initiation and maturation of autophagosomes.3, 4
In normal physiological conditions, autophagy proceeds at a modest basal level in virtually all cells to carry out homeostatic functions such as protein and organelle turnover. In response to such diverse stimuli as starvation, genotoxic chemicals and pathogen infection, autophagy process is induced for the maintenance of cellular metabolism or for the clearance of aggregated proteins, damaged organelles and intracellular pathogens, thus facilitating cell survival.5 Likewise, many anticancer agents are stressors requiring the autophagy response that assists the cancer cells to survive the crisis, and such cancer cells are in a state of ‘autophagy addiction’.6 The implication is that targeting autophagy addiction to block this important survival mechanism of cancer cells should be especially effective.6, 7
Pyrvinium is an FDA-approved antihelminthic drug for the treatment of enterobiasis.8 Recent studies have demonstrated that pyrvinium exerts potent toxicity against various cancer cell lines during glucose starvation and that pyrvinium decreases the viability of colon cancer cell lines and inhibits proliferation of myeloma cells.9, 10, 11
In our study, we demonstrate that pyrvinium potently inhibits autophagy by suppressing the transcription of autophagy genes and that targeting autophagy addiction by pyrvinium shows effective anticancer activity both in vitro and in vivo.
Results
Pyrvinium inhibits autophagosome formation
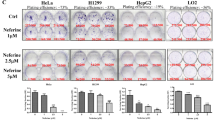
To determine whether pyrvinium affects autophagy in mammalian cell lines, we first used HeLa cells stably expressing GFP-LC3. We observed a decrease in GFP-LC3 puncta formation in pyrvinium-treated HeLa cells (Figure 1a), suggesting that autophagosome formation is inhibited by pyrvinium. To confirm these results, we assessed LC3 by immunoblot analysis, as the amount of LC3-II is correlated with the extent of autophagosome formation.12 LC3-II levels were reduced in HeLa cells treated with pyrvinium (Figure 1b), thus confirming that pyrvinium inhibited autophagosome formation. To investigate if these effects were restricted only to HeLa cells, we next used several different types of mammalian cell lines to analyze levels of LC3. In all cell lines studied, the LC3-II levels declined in response to pyrvinium treatment (Figure 1c), demonstrating a ubiquitous effect of pyrvinium on autophagosome formation. To analyze the effects of pyrvinium on the level of total LC3B protein, we used siAtg7 to abrogate the conversion of LC3-I to LC3-II, and pyrvinium was found to decrease the total LC3 protein levels in HeLa cells (Supplementary Figure 1). Furthermore, we found that the inhibiton of autophagy by pyrvinium was dose-dependent with an IC50 of 50 nM (Figure 1d). Finally, because pyrvinium blocks the basal autophagosome formation, it is possible that it could also block the induced autophagosome formation. Indeed, when cells were subjected to starvation in combination with pyrvinium, the number of autophagic vacuoles significantly decreased compared with that of starvation treatment alone (Figure 1e), indicating that pyrvinium could block starvation-induced autophagosome formation. Thus, the presence of pyrvinium inhibits both basal as well as starvation-induced autophagosome formation in mammalian cells.
Pyrvinium-induced autophagy arrest in mammalian cell lines. (a) GFP-LC3-expressing HeLa cells were treated with pyrvinium (30 nM) and vehicle control (1% DMSO) as indicated for 48 h. Quantitation on the right represents average GFP puncta per cell from three independent experiments±S.D.; 500 cells were analyzed per treatment condition. Scale bar, 20 μm. (b) GFP-LC3-expressing HeLa cells were treated as in a, and the cell lysates were analyzed by western blotting. (c) Four different cell lines as indicated were treated with pyrvinium (200 nM) and vehicle control (1% DMSO) for 24 h, and the cell lysates were analyzed by western blotting. (d) HeLa-GFP-LC3 cells were treated with vehicle control (1% DMSO) and indicated concentrations of pyrvinium for 48 h. The LC3 puncta were quantified. The image data were expressed as % of control vehicle treated cells from two independent experiments±S.D., 500 cells were analyzed per treatment condition. (e) HeLa cells were treated with pyrvinium (30 nM) and vehicle control (1% DMSO) in glucose-free media and then processed for EM analysis. The inset permits visualization of clearer autophagic vacuoles indicated by arrows. Data represent mean±S.E.M. from two independent experiments of at least 200 cells per sample. N, nucleus. Scale bar, 2 μm. Actin served as a loading control throughout. Band intensity was calculated using ImageJ software (National Institutes of Health, Bethesda, MD, USA), and the ratio of LC3-II/Actin expression was normalized and values are indicated. *P<0.05, **P<0.01 (unpaired t-test)
Pyrvinium decreases autophagic flux
LC3 analysis is now widely used to evaluate autophagy activity. However, the levels of LC3-II at a certain time point do not represent autophagic flux, therefore, it is important to measure the amount of LC3-II delivered to lysosomes by comparing LC3-II levels in the presence and absence of lysosomal inhibitors. Furthermore, LC3-II itself is an autophagy substrate, making interpretation of the results of LC3 immunoblot problematic, and as a result decreased LC3-II levels could be a reflection of autophagy enhancement and not de novo autophagy inhibition.13 To discriminate between these two possibilities, we analyzed autophagy in the presence of lysosomal inhibitors. Analysis of LC3-II levels in control cells showed increases in the amount of LC3-II when treated with NH4Cl (Figure 2a) or E64D and PEPS A (Supplementary Figure 2a), indicative of the basal level of autophagic flux; in contrast, pyrvinium caused a strong decline in LC3-II levels in the presence or absence of lysosomal inhibitors. These results demonstrated that pyrvinium blocked basal autophagic flux. To address whether pyrvinium inhibits stimuli-induced autophagic flux, diverse stimuli were used to stimulate autophagy.14 Stimulation of GFP-LC3-expressing HeLa cells with starvation or rapamycin (Figure 2b) or etoposide (Supplementary Figure 2b) led to increased LC3-II levels and an increased number of LC3 puncta (Figure 2c) that were further enhanced by lysosomal inhibitors NH4Cl and bafilomycin as expected. However, we also observed that pyrvinium caused decreases both in LC3-II levels and the numbers of LC3 puncta compared with that of stimuli treatment alone in the presence or absence of lysosomal inhibitors. Thus, pyrvinium also inhibits stimuli-induced autophagic flux. These results were further confirmed by measuring the rate of delivery of autophagosomes to lysosomes using a tandem monomeric RFP-GFP-tagged LC3.15 Starvation increased the numbers of both autophagosomes and autolysosomes, which were restored by pyrvinium to a level even lower than that of the control cells (Figure 2d), indicating that pyrvinium inhibited the starvation-induced autophagic flux. Together, the data presented in Figures 1 and 2 provide strong evidence that pyrvinium inhibits autophagy in vitro.
Pyrvinium attenuates both basal and stimuli-induced autophagic flux. (a) GFP-LC3-expressing HeLa and HEK293 cells were treated with pyrvinium (200 nM) and vehicle control (1% DMSO) in the presence or absence of NH4Cl (20 mM) for 48 h, and the cell lysates were analyzed by western blotting. Quantitation of immunoblot data in a on the right showing autophagic flux (change in LC3-II levels upon NH4Cl treatment). Values represent means normalized to actin from three independent experiments±S.E.M. (b) GFP-LC3-expressing HeLa cells in response to glucose starvation or rapamycin alone, or together with pyrvinium (200 nM) in the presence or absence of NH4Cl (20 mM) as indicated for 24 h, and the cell lysates were analyzed by western blotting. Quantitation of the immunoblot data in b on the right showing mean LC3-II levels±S.D. from NH4Cl-treated samples relative to untreated samples from three independent experiments and normalized to actin levels. (c) GFP-LC3-expressing HeLa cells were treated as in b. Quantitation shown on the right represents mean GFP puncta per cell (n=500) from two independent experiments±S.D. Scale bar, 20 μm. (d) HEK293 cells were transfected with RFP-GFP-LC3 plasmid encoding a dual-labeled LC3 protein. Cells were treated with pyrvinium (30 nM) and vehicle control (1% DMSO) for 48 h under glucose-free media. Representative images were shown as the average of vesicles per cell relative to the starvation treatment (%). For each cell, the number of AL (autolysosomes)=(mRFP-positive vesicles)/(GFP-negative vesicles); total autophagic vacuoles, mRFP-positive vesicles were enumerated. Data represent mean±S.D. for two independents experiments of at least 500 cells/sample. Scale bar, 20 μm. Star, starvation; Rapa, rapamycin; PP, pyrvinium. Actin served as a loading control throughout. Band intensity was calculated using ImageJ software, and the ratio of LC3-II/Actin expression was normalized and values are indicated. **P<0.01 (unpaired t-test)
Pyrvinium inhibits autophagy in vivo
To determine whether pyrvinium inhibits autophagy in vivo, we first investigated its effects on zebrafish. Studies in zebrafish larvae have demonstrated that alterations in LC3-II levels could be measured in the presence of NH4Cl, therefore allowing us to investigate in vivo changes in autophagic flux.16, 17 We observed an increase in LC3-II levels in zebrafish larvae treated with NH4Cl (Figure 3a). However, pyrvinium caused a significant decrease in LC3-II levels in the presence or absence of NH4Cl compared with the control, suggesting that pyrvinium inhibits autophagic flux in zebrafish larvae. We next used mice to examine whether pyrvinium inflicts the similar effects. Liver tissue was selected as an indicator because it is the best-characterized organ for changes in autophagic activity.16 Starvation strongly increased hepatic LC3-II levels, suggesting autophagy induction, but this increase was significantly reduced when treated with pyrvinium (Figure 3b). Thus, pyrvinium inhibited starvation-induced autophagy in mice. This result was further verified by immunohistochemical analysis of cytoplasmic LC3 puncta in liver specimens (Figure 3c). LC3 staining was generally weak in liver specimens from fed mice. However, in liver samples from starved mice, the presence of LC3 puncta was observed in the context of strong cytoplasmic staining, which was significantly recovered upon pyrvinium treatment. Interestingly, as predicted by our model, the increases in LC3-II levels in the intestine, heart and lung of starved mice were consistently reduced when treated with pyrvinium (Figure 3d), indicating that the total autophagy activity in starved mice might be inhibited by pyrvinium. Taken together, our studies suggested that pyrvinium inhibits autophagy in vivo.
Pyrvinium inhibits autophagy in vivo. (a) Zebrafish larvae were treated with pyrvinium (200 nM) and vehicle control (1% DMSO) in the presence or absence of NH4Cl (20 mM) for 24 h. Zebrafish larvae were then homogenized in lysis buffer and processed for western blotting. The panels showed autophagic flux (change in LC3-II levels upon NH4Cl treatment). Values represented means normalized to actin from three independent experiments±S.E.M. (b) Mice were divided into four groups. Two groups were treated with pyrvinium (10 mg/kg), whereas the other two groups were treated with vehicle control (5% CMC-Na) by oral gavage for 2 weeks. At the last 2 days, one of the control groups and one of the pyrvinium-treated groups were starved for 48 h. After killing, the liver was dissected and homogenized in lysis buffer before being analyzed by western blotting. The graph represented the mean results from five mice per group with control set to 100%. Error bars represent S.E.M. (c) Immunohistochemical detection of cytoplasmic LC3 puncta in liver specimens from mice treated as in b. Representative images were provided as indicated. Scale bars, 20 μm. Arrowheads depicted LC3 dots. (d) Intestine, heart and lung tissues from starved mice treated with pyrvinium or vehicle control (5% CMC-Na) were isolated and analyzed by western blotting. Actin served as a loading control throughout. Band intensity was calculated using ImageJ software, and the ratio of LC3-II/actin expression was normalized and values are indicated. NS, not significant. **P<0.01 (unpaired t-test)
Regulation of autophagy by pyrvinium is AMPK/mTOR independent
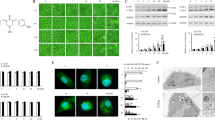
Pyrvinium is a quinoline-derived cyanine dye; the red fluorescence of pyrvinium can be observed by using fluorescent microscopy.10 We found that pyrvinium formed dot-like aggregates (Supplementary Figure 3a). Pyrvinium aggregates did not colocalize with the lysosome marker LAMP-1 or autophagosome marker LC3, but with mitochondria markers cytochrome-c and V-DAC or MitoTracker fluorescence signal (Supplementary Figure 3b), these data suggest that pyrvinium has a preferential localization in mitochondria. In the presence of carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP), a proton ionophore that abolishes the electrochemical gradient, a very small amount of pyrvinium aggregates was detected (Supplementary Figure 3c). Moreover, similar results were obtained by incubating the HEK293 cells in a high K+ buffer that dissipates the plasma membrane potential (Supplementary Figure 3c). These results suggested that the formation of pyrvinium aggregates within mitochondria is dependent on the presence of a negative sink inside mitochondria created by proton pumps. It has recently been reported that pyrvinium inhibits the mitochondrial respiratory chain complex I activity, leading to decreased ATP production in human myeloma/erythroleukemia cells.11 We then analyzed the ATP levels in HeLa and HEK293 cells treated with pyrvinium. Cellular ATP levels were significantly decreased by pyrvinium in a concentration-dependent manner (Supplementary Figure 3d). AMPK is activated by the phosphorylation of the a-subunit of AMPK after an increase in the intracellular AMP/ATP ratio.5 Decreased cellular ATP levels by pyrvinium activated AMPK in Hela and HEK293 cells (Supplementary Figure 3e). mTOR has emerged as a key negative regulator of autophagy, and the activity of mTOR can be regulated by AMPK.18 Activation of AMPK by pyrvinium inversely correlated with inhibition of the mTOR (Supplementary Figure 3e). The inhibition of the mTOR pathway was further confirmed by the blocked phosphorylation status of both ribosomal protein subunit S6 and eIF4E-binding protein (4EBP1). Interestingly, we found that exogenous ATP added to the cells after the addition of the pyrvinium restored the activity of AMPK and mTOR (Supplementary Figure 3f), demonstrating that pyrvinium influenced the AMPK/mTOR pathway via decreasing ATP levels. Moreover, compound C, an inhibitor of AMPK, did not influence the effect of pyrvinium on LC3-II accumulation (Supplementary Figure 3g), suggesting that inhibition of autophagy by pyrvinium is AMPK independent. As mentioned above, even in the presence of rapamycin, an inhibitor of mTOR, pyrvinium suppresses autophagy (Figures 2b and c). Thus, inhibition of autophagy by pyrvinium is AMPK/mTOR independent.
Pyrvinium reduces the transcription of autophagy-associated genes
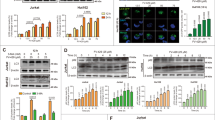
According to our observations, we speculated that the presence of an mTOR-independent mechanism was responsible for the control of autophagy by pyrvinium. To address this hypothesis, we next examined protein levels of the class 3 phosphatidylinositol kinase Vps34/p150/Beclin1/Atg14 complex 1 that regulates autophagy, independent of mTOR function.19 Treatment with pyrvinium reduced the levels of Beclin1, Vps34, Atg14 and p150 in HeLa and HEK293 cells, and the effect of pyrvinium on the levels of Vps34 complex was strongly correlated with that of LC3-II (Figure 4a), indicating that pyrvinium-mediated inhibition of autophagy was associated with reduction in Vps34 complex levels. To address how pyrvinium reduces the levels of Vps34 complex, we conducted a time course study in HeLa cells treated with pyrvinium and found that the appearance of decreased levels of Vps34 complex relatively needed longer time periods (Figure 4b, Supplementary Figure 4a); this raises the possibility that pyrvinium does not promote the degradation of Vps34 complex. Therefore, we next treated HeLa cells with pyrvinium in the presence of cycloheximide (CHX). The addition of pyrvinium with CHX did not reduce the levels of Vps34 complexes compared with that of CHX treatment alone (Figure 4c), suggesting that pyrvinium does not promote the degradation but impairs the production of Vps34 complexes. In an attempt to understand whether pyrvinium regulates the transcription of Vps34 complex components, we used real-time PCR to measure the mRNA levels in HeLa cells. The results showed that pyrvinium reduced the Vps34, Beclin1, p150 and Atg14 mRNA levels in a dose-dependent manner (Figure 4d). Similar inhibitory effects were observed on LC3B, Atg7 and Atg5 (Supplementary Figure 4b). However, in the presence of actinomycin, an inhibitor of DNA transcription, pyrvinium did not further decrease the mRNA levels of Beclin1 and LC3B (Supplementary Figure 4c), suggesting that pyrvinium reduces the transcription of autophagy-associated genes. In previous studies, pyrvinium had been shown to specifically inhibit the transcription of target genes of Wnt signaling; in contrast, pyrvinium had no discernible effect on the transcriptional responses of four other major signaling pathways (TGFα, BMP4, IL-4 and Notch).10 In this study, we found that the mRNA and protein levels of some metabolic associated genes (PKM2, PGAM1and GSK3β) and exogenous GFP expression did not change upon treatment of pyrvinium (Supplementary Figures 4d and e), suggesting pyrvinium as a negative transcriptional regulator of autophagy.
Pyrvinium inhibits the transcription of some autophagy genes. (a) HeLa and HEK293 cells were treated with pyrvinium (200 nM) and vehicle control (1% DMSO) for 24 h, and the cell lysates were analyzed by western blotting. (b) HeLa cells were treated with pyrvinium (200 nM) and vehicle control (1% DMSO) for the indicated periods of time, and the cell lysates were analyzed by western blotting. (c) HeLa cells were treated with pyrvinium (200 nM) for the 24 h in the presence of CHX (50 μg/ml), and the cell lysates were analyzed by western blotting. (d) Hela cells were treated with indicated concentration of pyrvinium for 24 h, and the mRNA expression of the Beclin1, Vps34, Atg14 and p150 were analyzed by real-time PCR. All values were normalized to the mRNA level of actin. All error bars indicate STD
Pyrvinium combined with autophagy stimuli sensitizes cancer cells to apoptosis
To further characterize the biological effects of pyrvinium at the cellular level, we analyzed the cytotoxicity of pyrvinium in selected human cancer cell lines. Pyrvinium inhibited the growth (Figure 5a) and clone formation (Figure 5b) of HeLa and HCT116 cells in a dose-dependent manner, but did not significantly induce apoptotic cell death (Supplementary Figures 5a and b). These results indicated that the main growth inhibition induced by pyrvinium was not due to induction of apoptosis. It has been reported that the anticancer activity of pyrvinium shows preferential cytotoxicity during glucose starvation.9 Starvation is the most well-known conventional inducer of autophagy. To determine whether other autophagy stimuli could also enhance the cytotoxicity of pyrvinium against cancer cells, we treated HeLa cells with pyrvinium in the presence of glucose starvation, etoposide or Pseudomonas aeruginosa.14, 20 All three combined treatments caused dramatically enhanced cell death compared with pyrvinium treatment alone (Figure 5c). Cells treated with pyrvinium in combination with autophagy stimuli showed apoptotic morphology (Figure 5d), characteristic caspase 3 and PARP cleavage (Figure 5e), and increases in the number of terminal transferase dUTP nick-end labeling (TUNEL)-positive cells (Supplementary Figure 5c). Similar results were obtained with MCF7 and PANC1 cells (data not shown). LC3 immunoblot studies further confirmed that autophagy was triggered by autophagy stimuli and that the induced autophagy was inhibited by the addition of pyrvinium (Figure 5f). To test whether enhanced cell death is due to targeted autophagy addiction, we treated HeLa cells with commonly used autophagy inhibition agents under starvation condition (Supplementary Figures 5d and e). Similar to pyrvinium treatment, all three autophagy inhibitors showed increased cytotoxicity under starvation condition compared with that of normal growth conditions, suggesting that targeting autophagy addiction promotes cancer cell death. We next examined the cell viability of HeLa cells pretreated with 3-MA or siBeclin1 and then administered with pyrvinium under starvation condition. Pretreatment of cells with 3-MA or siBeclin1 partially protected cells from pyrvinium-induced cell death under glucose starvation (Figures 5g and h), suggesting that targeted autophagy addiction is involved in pyrvinium-mediated cytotoxicity. Thus, our data suggest that pyrvinium sensitizes cancer cells to apoptosis in combination with autophagy stimuli, and this effect can at least be partially attributed to targeting the autophagy addiction on which cell depends for survival under harsh environmental conditions.
Pyrvinium combined with autophagy stimuli sensitizes cancer cells to apoptosis. (a) HeLa and HCT116 cells were treated with vehicle control (1% DMSO) and indicated concentration of pyrvinium for 2 days, and the cell proliferation was determined by WST-8 assay. (b) The colony formation assay was performed in HeLa and HCT116 cells treated with vehicle control (1% DMSO) or indicated concentration of pyrvinium. (c–e) HeLa cells were treated with vehicle control (1% DMSO) or indicated concentration of pyrvinium for 24 h before subjected to autophagic induction by starvation for 12 h or etoposide (10 μm) for 24 h or P. aeruginosa infection for 4 h. (c) Cell viability was determined by MTT assay. (d) Representative morphology imaged using a phase contrast microscope. (e) The apoptotic pathway was analyzed by western blotting using the indicated antibodies. (f) HeLa cells were treated as in c, and the cell lysates were analyzed by western blotting. (g and h) HeLa cells were pretreated with siBeclin1 (g) or 3-MA (h) and then administered with pyrvinium under starvation condition for 24 h, and the cell viability was determined by MTT assay. Survival values (%) are indicated. Actin served as a loading control throughout. Band intensity was calculated using ImageJ software, and the ratio of LC3-II/Actin expression was normalized and values are indicated. All error bars indicate STD.
Pyrvinium combined with 2-deoxy-D-glucose displays more potent anticancer activity in vivo
To examine whether the combination therapy of a chemotherapeutic agent to induce autophagy and pyrvinium to target the aroused autophagy addiction would result in greater efficacy than either agent alone, we used the autophagy inducer 2-deoxy-D-glucose (2-DG) in combination with pyrvinium in a xenograft mouse model. 2-DG was reported to induce autophagy by therapeutic starvation.21 We first tested the efficacy of combination treatments in vitro. Similar to treatment of pyrvinium under starvation condition, the combination of pyrvinium with 2-DG was more toxic against 4T1 cancer cells than either treatment alone (Supplementary Figures 6a and b). As predicted, 2-DG-induced autophagy was inhibited by pyrvinium in 4T1 cells (Supplementary Figure 6c). Pyrvinium, in combination with 2-DG, was clearly more effective in reducing tumor volume and weight as compared with either treatment alone (Figures 6a–c). Immunohistochemical analysis of cytoplasmic LC3 puncta in tumor samples confirmed that the 2-DG-triggered autophagy was inhibited by pyrvinium (Figure 6d), suggesting that pyrvinium targets autophagy addiction caused by 2-DG in these tumors. Massive activation of caspase 3 was observed in tumor samples upon combination treatment (Figure 6e). In the experiment where treatment began when tumors were 600 mm3, the combination chemotherapy could also induce apoptosis, leading to a decrease of tumor size (Figure 6f, Supplementary Figure 7a). Thus, the combination chemotherapy with pyrvinium and 2-DG showed more potent growth suppressive effects in vivo. Moreover, the combination treatment showed minimal toxic effects in normal animals (Supplementary Figures 7b–g). The modest effects on body weights and liver size appear to be related to the reported adverse effects of orally administered pyrvinium.22
Effect of pyrvinium and 2-DG, alone and in combination, on growth of 4T1 tumor xenografts. 4T1 tumor-bearing mice were treated with pyrvinium and 2-DG, alone and in combination, at 4 weeks after implantation. (a) Representative picture of formed tumors. (b) Growth curve of tumor volumes. (c) Tumor weight. (d) Immunohistochemical detection of cytoplasmic LC3 puncta in tumor specimens. Representative images were provided as indicated. Scale bars, 20 μm. Arrowheads depicted LC3 dots. (e) Immunohistochemical detection of cleaved caspase 3 in tumor specimens. Representative images were provided as indicated. Scale bars, 20 μm. (f) Growth curve of tumor volumes started from 600 mm3. **P<0.001 (unpaired t-test)
Discussion
In this study, we identify pyrvinium as a potent small molecule inhibitor of autophagy with an IC50 of 50 nM. Using zebrafish and mice to assess the effects of pyrvinium on autophagy in vivo, we found that pyrvinium decreases their autophagy levels.
We observed that pyrvinium localized to mitochondria and identified pyrvinium as a lipophilic cation. Lipophilic cations such as rhodamine-123 have been used to monitor mitochondrial membrane potential in a single living cell.23 Thus, because of its innate fluorescent properties and low toxicities, pyrvinium could be used as a new probe for monitoring mitochondrial membrane potential. Pyrvinium accumulates preferentially in mitochondria and disrupts respiratory chain complex I, leading to decreased ATP production, which causes AMPK activation and downstream inhibition of mTOR. The inhibitory function of mTOR in autophagy is well established.24 However, recent screens of FDA-approved drugs have identified numerous mTOR-independent autophagy modulators.25 We show that pyrvinium decreases Vps34 complex levels by inhibiting the transcription of its components. In recent years there has been a rapid increase in reports on transcriptional regulation of autophagy.26, 27 In a previous study, pyrvinium was shown to selectively bind to the minor groove of AT-rich DNA regions.28 Whether this property of pyrvinium is associated with its inhibitory effect on the transcription of autophagy genes needs further investigation.
We demonstrate that pyrvinium inhibits proliferation but does not cause significant apoptotic cancer cell death under normal physiological conditions; however, massive cell death occurs in response to pyrvinium when subjected to worse conditions such as starvation, genotoxic insult or pathogen infection. We speculate that under these conditions, cancer cells are in a state of autophagy addiction requiring the autophagic process for promoting cell survival, whereas this important survival pathway can be blocked by pyrvinium, thus generating the observed phenomenon of massive cancer cell death. Using chloroquine (CQ), 3-MA and siBeclin1 to inhibit autophagy, elevated numbers of cancer cells die under starvation conditions compared with normal conditions. These observations support a role of targeting autophagy addiction by pyrvinium treatment to promote cancer cell death. However, except for its function as an autophagy inhibitor, pyrvinium has multiple other roles.10, 29, 30 It has also been shown that the suppression of unfolded protein responses is involved in pyrvinium-mediated cytotoxicity under glucose starvation29; whether inhibited Wnt signaling and androgen receptor activity could also contribute to these effect is unclear. Although future research studies are required to provide more direct evidence for the above conclusions, there is no doubt that autophagy inhibition by pyrvinium partially contributes to cancer cell death in the context of targeting autophagy addiction.
The notion of autophagy modulation as a potential therapeutic target for cancer has been recently discussed in the literature.31, 32, 33, 34, 35 Accordingly, there are several Phase I/II clinical trials in progress using the lysosome inhibitors CQ or hydroxychloroquine in combination with chemotherapeutic agents for the treatment of some tumors.34 As pyrvinium is determined as a potent autophagy inhibitor, we studied its use as a cancer treatment in vivo to target autophagy addiction. To create the state of autophagy addiction in cancer, 2-DG was selected because it induces autophagy by therapeutic starvation.21 Moreover, 2-DG appears to be safe when administered to animals.36 Xenograft experiments demonstrated that pyrvinium combination therapy with 2-DG shows significantly enhanced anticancer efficacy in vivo, supporting our hypothesis of targeting autophagy addiction by pyrvinium in cancer. In addition, the combination therapy, which exhibits minimal toxicity in normal animals, appears to be a promising cancer-treatment candidate.
As discussed earlier, many open questions and controversies remain regarding autophagy as a target of pyrvinium in cancer therapy. Although the available data support the concept that cancer cells undergoing increased autophagy die in response to pyrvinium, no direct evidence have unequivocally demonstrated that the synergistic inhibition of 4T1 tumor growth by the combination of pyrvinium and 2-DG arises specifically from autophagy inhibition. However, given the strong potential of pyrvinium as a lead compound for developing specific and potent autophagy inhibitor, screening out the pyrvinium derivatives without autophagy-independent functions would help us to further elucidate the existing paradigms that dictate our current understanding of autophagy as a target in cancer therapy.
Materials and Methods
Cell culture and treatment
HeLa, HeLa-GFP-LC3, HEK293, PANC1, HCT116 and MEF cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA) and 4T1 cells were cultured in RPMI1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA), penicillin (107 U/l) and streptomycin (10 mg/l) at 37 °C in a humidified chamber containing 5% CO2. For glucose starvation experiments, cells were cultured in DMEM only without glucose (GIBCO, Grand Island, NY, USA).
Transfection and oligonucleotides
Transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. siRNA for Beclin1 (sense 5′-GGAGCCAUUUAUUGAAACUTT-3′ and antisense 5′-GUUUCAAUAAAUGGCUCCTT-3′) or Atg7 (sense 5′-GCCTGCTGAGGAGCTCTCCAT-3′ and antisense 5′-AAGGAAGAGCTGTGACTCC-3′) were synthesized by Genepharma (Shanghai, China).
Antibodies and reagents
Rabbit polyclonal antibody anti-LC3 was from Novus (Littleton, CO, USA). Rabbit polyclonal antibody anti-Vps34 and rabbit monoclonal antibody anti-p150 and anti-Cleaved caspase 3 were from Cell Signaling (Beverly, MA, USA). Rabbit polyclonal antibody anti-Atg14, anti-Beclin1, anti-UVRAG, anti-Atg5, anti-Atg7, anti-PKM2, anti-PGAM1 and anti-GSK3β were from MBL (Nagoya, Japan). Rabbit polyclonal antibody anti-AMPKα, anti-phospho-AMPKα, anti-mTOR, anti-phospho-mTOR, mouse monoclonal antibody anti-caspase 3, anti-PARP and anti-GFP were from Millipore (Bedford, MA, USA). Rabbit polyclonal antibody anti-4EBP1, anti-phospho-4EBP1, anti-S6K, anti-phospho-S6K and mouse monoclonal antibody anti-β-actin were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Pyrvinium pamoate was from US Pharmacopeia (Rockville, MD, USA). Pepstatin A, E64d, rapamycin, CHX, etoposide, CQ, bafilomycin, FCCP, ATP, MitoTracker green, compound C and 2-DG were from Sigma (St. Louis, MO, USA).
Tissue and cell lysates and western blotting
Tissue samples were homogenized in lysis buffer (50 mM Tris-base, 1.0 mM EDTA, 150 mM NaCl, 0.1% SDS, 1% TritonX-100, 1% sodium deoxycholate and 1 mM phenylmethylsulfonyl fluoride). After centrifugation at 5000 × g for 5 min, the supernatant fractions were collected. Cells were solubilized in lysis buffer for 1 h at 4 °C and lysates were centrifuged at 5000 × g for 20 min. Total homogenates were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Amersham Biosciences, Piscataway, NJ, USA) as previously described.37 Upon incubation with primary and secondary antibodies, immunoreactive bands were detected by ECL according to the manufacturer’s instructions.
Immunohistochemistry
Immunohistochemistry was performed using the Dako EnVision Systems (Dako Cytomation GmbH, Hamburg, Germany).38 Consecutive paraffin wax-embedded tissue sections (3–5 μm) were dewaxed and rehydrated. Antigen retrieval was performed by pretreatment of the slides in citrate buffer (pH 6.0) in a microwave oven for 12 min. Thereafter, slides were cooled to room temperature in deionized water for 5 min. Endogenous peroxidase activity was quenched by incubating the slides in methanol containing 0.6% hydrogen peroxide followed by washing in deionized water for 3 min after which the sections were incubated for 1 h at room temperature with normal goat serum and subsequently incubated at 4 °C overnight with the primary antibodies rabbit anti-LC3 (diluted 1 : 200) or anti-Cleaved caspase 3 (diluted 1 : 500). Next, the sections were rinsed with washing buffer (TBS with 0.1% bovine serum albumin) and incubated with horseradish peroxidase-linked goat anti-rabbit antibodies followed by reaction with diaminobenzidine and counterstaining with Mayer’s hematoxylin. Immunostaining was detected using 3,3′-diaminobenzidine substrate solution (Dako Cytomation GmbH) according to the manufacturer’s instructions.
Electron microscopy
Pretreated cells were fixed in 0.1% glutaraldehyde, dehydrated and embedded. Ultrathin sections (70 nm) were cut, stained and analyzed by a Philips EM420 electron microscopy (Philips instruments, Inc., Mahwah, NJ, USA).
ATP measurement
Cells were incubated with pyrvinium for 24 h, and the cell suspension was transferred to a 96-well microplate (BD Biosciences, San Jose, CA, USA). Cellular ATP levels were examined using an ATP Assay Kit (Invitrogen).
Real-time PCR
Total cellular RNA was isolated with TRIzol (Invitrogen). cDNA synthesis was performed using Improm-II Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The Vps34 mRNA was assayed using the primers sense: 5′-GGACCTTCTGACCACGAT-3′ and antisense: 5′-GCAACAGCATAACGCCTC-3′; the Beclin1 mRNA with the primers sense: 5′-CGTGGAATGGAATGAGAT-3′ and antisense: 5′-GGTCAAACTTGTTGTCCC-3′; the ATG14L mRNA with the primers sense: 5′-GAGCGGCGATTTCGTCTACT-3′ and antisense: 5′-CTGAAGACACATCTGCGGGG-3′; the p150 mRNA with the primers sense: 5′-AGAAGGGCAAGCTACAGA-3′ and antisense: 5′-AATTCAGGGCAGAGGAG-3′; the LC3B mRNA with the primers sense: 5′-AGACCTTCAAGCAGCGCCG-3′ and antisense: 5′-ACACTGACAATTTCATCCCG-3′; the Atg5 mRNA with the primers sense: 5′-AAGCAACTCTGGATGGGA TT-3′ and antisense: 5′-GCAGCCACAGGACGAAAC-3′; the Atg7 mRNA with the primers sense: 5′-TTTGCTATCCTGCCCTCT-3′ and antisense: 5′-TGCCTCCTTTCTGGTTCT-3′; and the β-actin mRNA with the primers sense: 5′-CACGATGGAGGGGCCGGACTCATC-3′ and antisense:5′-TAAAGACCTCTATGCCAACAC GT-3′. Reverse transcription of purified RNA was performed using oligo (dT) primer. The quantification of gene transcripts was performed by real-time PCR using SYBR Green I dye (Invitrogen). All values of genes were normalized to the level of β-actin mRNA.
Bacteria strains
P. aeruginosa strain PAO1 wild-type (WT) was a gift from Stephen Lory (Harvard Medical School, Boston, MA). Bacteria were grown overnight in Luria–Bertani (LB) broth at 37 °C with vigorous shaking. The next day, the bacteria were pelleted by centrifugation at 8000 × g and resuspended in 10 ml of fresh LB broth, in which they were allowed to grow until the mid-logarithmic phase. Thereafter, the optical density (OD) at 600 nm was measured, and the density was adjusted to 0.25 OD (0.1 OD=1 × 108 cells/ml). Cells were washed once with PBS after overnight culture in serum-containing medium and changed to serum-free and antibiotic-free medium immediately before infection.
Cell proliferation, viability and apoptotic cell death measurement
Cells were seeded onto 96-well microplates and treated with pyrvinium. After several days of incubation, cell proliferation was evaluated by the WST-8 colorimetric assay using a Cell Count Reagent SF reagent (Nacalai Tesque, Kyoto, Japan). Cell viability was analyzed by MTT assay as previously described.39 Apoptosis was measured by TUNEL assay as previously described.40
Zebrafish and mouse experiments
Autophagy assays in these animals were performed as previously described.17 The Institutional Animal Care and Treatment Committee of Sichuan University approved all studies herein. Briefly, after the determination of maximum tolerated concentration of pyrvinium in larval zebrafish, endogenous LC3-II assays were performed at the following concentrations: NH4Cl at 100 mM and pyrvinium at 200 nM for 24 h. WT larvae (n=30 per treatment group) at 2 d.p.f. were exposed to pyrvinium or embryo medium (untreated control) for 20 h before compound replenishment and the addition of NH4Cl. Larvae were incubated in compounds for a further 4 h, homogenized in lysis buffer, processed for western blotting as described above. For mouse experiments, 20 healthy female mice (BALB/c, 6–8 weeks of age, nonfertile and 18–20 g each) were used at the start of the study. The mice were divided into four groups. After 2 weeks of acclimatization, two groups were treated with pyrvinium (10 mg/kg), whereas the other two groups were treated with vehicle control (5% CMC-Na) by oral gavage for 2 weeks. At the last 2 days, one of the control groups and one of the pyrvinium-treated groups were starved for 48 h. After killing, the liver was dissected and homogenized in lysis buffer before being analyzed by western blotting.
Tumor xenograft model
Healthy female mice (BALB/c, 6–8 weeks of age, nonfertile and 18–20 g each) were injected subcutaneously with 4T1 cells (one million cells per mouse). When tumors were approximately 50 or 600 mm3 in size, the animals were pair-matched into four groups (eight mice per group) as follows: 5% CMC-Na-treated control, 2-DG alone, pyrvinium alone and pyrvinium+2-DG. At day 0, the 2-DG alone and pyrvinium+2-DG groups received 0.2 ml of 2-DG i.p. at 150 mg/ml (1000 mg/kg), which was repeated every 2 days for the duration of the experiment. On day 1, the pyrvinium and pyrvinium+2-DG groups received 0.2 ml of pyrvinium i.g. at 1 mg/ml (10 mg/kg), which was repeated once a day. This experiment was then repeated (n=3/group) but was delayed until the tumors were approximately 1000 mm3. Tumor measurements were taken by caliper every 3 days and converted to tumor volume by using the formula W × L2/2. At the time of killing, tumors were excised, pictured and weighed.
Toxicity assessment
BALB/c female mice, six animals per group, were dosed as described in tumor xenograft model experiment. Body weight was determined every 3 days until the end of the study. At the time of killing, tissues were harvested, weighted and prepared for H&E staining.
Abbreviations
- ATG:
-
autophagy-related gene
- TUNEL:
-
terminal transferase dUTP nick-end labeling
- FCCP:
-
carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone
- CQ:
-
chloroquine
- AMPK:
-
AMP-activated protein kinase
- 2-DG:
-
2-deoxy-D-glucose
- mTOR:
-
mammalian target of rapamycin
- 4EBP1:
-
eIF4E-binding protein
- CHX:
-
cycloheximide
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- WT:
-
wild-type
- LB:
-
Luria–Bertani
- OD:
-
optical density
References
Behrends C, Sowa ME, Gygi SP, Harper JW . Network organization of the human autophagy system. Nature 2010; 466: 68–76.
Kim J, Kundu M, Viollet B, Guan KL . AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13: 132–141.
Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011; 147: 223–234.
Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol 2010; 12: 665–675.
Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T et al. The role of autophagy during the early neonatal starvation period. Nature 2004; 432: 1032–1036.
Garber K . Inducing indigestion companies embrace autophagy inhibitors. J Natl Cancer Inst 2011; 103: 708–710.
Mancias JD, Kimmelman AC . Targeting autophagy addiction in cancer. Oncotarget 2011; 2: 1302–1306.
Downey AS, Chong CR, Graczyk TK, Sullivan DJ . Efficacy of pyrvinium pamoate against Cryptosporidium parvum infection in vitro and in a neonatal mouse model. Antimicrob Agents Chemother 2008; 52: 3106–3112.
Esumi H, Lu J, Kurashima Y, Hanaoka T . Antitumor activity of pyrvinium pamoate, 6-(dimethylamino)-2-[2-(2,5-dimethyl-1-phenyl-1H-pyrrol-3-yl)ethenyl]-1-methyl-quinolinium pamoate salt, showing preferential cytotoxicity during glucose starvation. Cancer Sci 2004; 95: 685–690.
Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat Chem Biol 2010; 6: 829–836.
Harada Y, Ishii I, Hatake K, Kasahara K . Pyrvinium pamoate inhibits proliferation of myeloma/erythroleukemia cells by suppressing mitochondrial respiratory complex I and STAT3. Cancer Lett 2012; 19: 83–88.
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19: 5720–5728.
Mizushima N, Yoshimori T . How to interpret LC3 immunoblotting. Autophagy 2007; 3: 542–545.
Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta H, Kanaseki T et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009; 461: 654–658.
Joubert PE, Werneke SW, de la Calle C, Guivel-Benhassine F, Giodini A, Peduto L et al. Chikungunya virus–induced autophagy delays caspase-dependent cell death. J Exp Med 2012; 209: 1029–1047.
Underwood BR, Imarisio S, Fleming A, Rose C, Krishna G, Heard P et al. Anti oxidants can inhibit basal autophagy and enhance neurodegeneration in models of polyglutamine disease. Hum Mol Genet 2010; 19: 3413–3429.
Hart PD, Young MR . Ammonium chloride, an inhibitor of phagosome–lysosome fusion in macrophages, concurrently induces phagosome–endosome fusion, and opens a novel pathway: studies of a pathogenic mycobacterium and a nonpathogenic yeast. J Exp Med 1991; 174: 881–889.
Hardie DG . AMPK and Raptor: matching cell growth to energy supply. Mol Cell 2008; 30: 263–265.
Yamada E, Bastie CC, Koga H, Wang Y, Cuervo AM, Pessin JE . Mouse skeletal muscle fiber-type-specific macroautophagy and muscle wasting are regulated by a Fyn/STAT3/Vps34 signaling pathway. Cell Rep 2012; 1: 557–569.
Yuan K, Huang C, Fox J, Laturnus D, Carlson E, Zhang B et al. Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. J Cell Sci 2012; 125: 507–515.
DiPaola RS, Dvorzhinski D, Thalasila A, Garikapaty V, Doram D, May M et al. Therapeutic starvation and autophagy in prostate cancer: a new paradigm for targeting metabolism in cancer therapy. Prostate 2008; 68: 1743–1752.
Tomitsuka E, Kita K, Esumi H . An anticancer agent, pyrvinium pamoate inhibits the NADH-fumarate reductase system–a unique mitochondrial energy metabolism in tumour microenvironments. J Biochem 2012; 152: 171–183.
Smiley ST, Reers M, Mottola-Hartshorn C, Lin M, Chen A, Smith TW et al. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci USA 1991; 88: 3671–3675.
Klionsky DJ, Emr SD . Autophagy as a regulated pathway of cellular degradation. Science 2000; 290: 1717–1721.
Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH et al. Novel targets for Huntingtons disease in an mTOR-independent autophagy pathway. Nat Chem Biol 2008; 4: 295–305.
Polager S, Ofir M, Ginsberg D . E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 2008; 27: 4860–4864.
Toepfer N, Childress C, Parikh A, Rukstalis D, Yang W . Atorvastatin induces autophagy in prostate cancer PC3 cells through activation of LC3 transcription. Cancer Biol Ther 2011; 12: 691–699.
Stockert JC, Trigoso CI, LIorente AR, Del Castillo P . DNA fluorescence induced pyrvinium binding by polymethine cation. Histochem J 1991; 23: 548–552.
Yu DH, Macdonald J, Liu G, Lee AS, Ly M, Davis T et al. Pyrvinium targets the unfolded protein response to hypoglycemia and its anti-tumor activity is enhanced by combination therapy. PLoS One 2008; 3: e3951.
Jones JO, BoIton EC, Huang Y, Feau C, Guy RK, Yamamoto KR et al. Non-competitive androgen receptor inhibition in vitro and in vivo. Proc Natl Acad Sci USA 2009; 106: 7233–7238.
Yang ZJ, Chee CE, Huang S, Sinicrope FA . The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011; 10: 1533–1541.
Janku F, McConkey DJ, Hong DS, Kurzrock R . Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol 2011; 8: 528–539.
Mah LY, Ryan KM . Autophagy and cancer. Cold Spring Harb Perspect Biol 2012; 4: a008821.
Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 2011; 17: 654–666.
Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI et al. Autophagy inhibiton enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 2007; 117: 326–336.
Landau BR, Lubs HA . Animal responses to 2-deoxy-D-glucose administration. Proc Soc Exp Biol Med 1958; 99: 124–127.
Lei Y, Huang K, Gao C, Lau QC, Pan H, Xie K et al. Proteomics identification of ITGB3 as a key regulator in reactive oxygen species-induced migration and invasion of colorectal cancer cells. Mol Cell Proteomic 2011; 10: M110 005397.
Liu R, Li Z, Bai S, Zhang H, Tang M, Lei Y et al. Mechanism of cancer cell adaptation to metabolic stress: proteomics identification of a novel thyroid hormone-mediated gastric carcinogenic signaling pathway. Mol Cell Proteomic 2009; 8: 70–85.
Yuan K, Huang C, Fox J, Gaid M, Weaver A, Li G et al. Elevated inflammatory response in caveolin-1-deficient mice with Pseudomonas aeruginosa infection is mediated by STAT3 protein and nuclear factor B (NF-κB). J Biol Chem 2011; 286: 21814–21825.
Wang K, Liu R, Li J, Mao J, Lei Y, Wu J et al. Quercetin induces protective autophagy in gastric cancer cells: involvement of Akt-mTOR- and hypoxia-induced factor 1α-mediated signaling. Autophagy 2011; 7: 966–978.
Acknowledgements
This work was supported by grants from the National 973 Basic Research Program of China (2013CB911300, 2011CB910703 and 2012CB518900), the National Science and Technology Major Project (2011ZX09302-001-01 and 2012ZX09501001-003), Doctoral fund from Chinese MOE (20120181110024) and Chinese NSFC (81225015, 81072022 and 81172173).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by H-U Simon
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Deng, L., Lei, Y., Liu, R. et al. Pyrvinium targets autophagy addiction to promote cancer cell death. Cell Death Dis 4, e614 (2013). https://doi.org/10.1038/cddis.2013.142
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.142
Keywords
This article is cited by
-
Predictive value of CCL2 in the prognosis and immunotherapy response of glioblastoma multiforme
BMC Genomics (2023)
-
Retinoic acid regulates endometriotic stromal cell growth through upregulation of Beclin1
Archives of Gynecology and Obstetrics (2018)
-
Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets
Nature Communications (2017)
-
Enhanced Autophagy Contributes to Protective Effects of GM1 Ganglioside Against Aβ1-42-Induced Neurotoxicity and Cognitive Deficits
Neurochemical Research (2017)
-
RETRACTED ARTICLE: Effect of pantoprazole to enhance activity of docetaxel against human tumour xenografts by inhibiting autophagy
British Journal of Cancer (2015)