Abstract
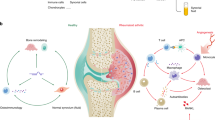
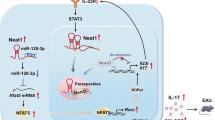
microRNAs are a novel group of small, conserved, non-coding RNA molecules that are present in all species. These molecules post-transcriptionally regulate gene expression by targeting mRNAs for degradation or by repressing the translation of the mRNAs. A good understanding of miRNA-mediated gene regulation is critical to gain a comprehensive view of many physiological processes and disease states. Emerging evidence demonstrates that miRNAs play an important role in the differentiation and function of the adaptive immune system. This review provides an overview of the diverse functions of miRNAs in modulating immune responses and in immune cell development, particularly the development of Th17 cells, and explores the involvement of miRNAs in several autoimmune diseases including multiple sclerosis (MS), rheumatoid arthritis (RA), inflammatory bowel disease (IBD) and diabetes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Dong C . Differentiation and function of pro-inflammatory Th17 cells . Microbes Infect 2009 ; 11 : 584 – 588 .
Miossec P, Korn T, Kuchroo VK . Interleukin-17 and type 17 helper T cells . N Engl J Med 2009 ; 361 : 888 – 898 .
Ambros V . The functions of animal microRNAs . Nature 2004 ; 431 : 350 – 355 .
Bartel DP, Chen CZ . Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs . Nat Rev Genet 2004 ; 5 : 396 – 400 .
Bushati N, Cohen SM . microRNA functions . Annu Rev Cell Dev Biol 2007 ; 23 : 175 – 205 .
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets . Cell 2005 ; 120 : 15 – 20 .
Xiao C, Rajewsky K . MicroRNA control in the immune system: basic principles . Cell 2009 ; 136 : 26 – 36 .
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A . Identification of mammalian microRNA host genes and transcription units . Genome Res 2004 ; 14 : 1902 – 1910 .
Cai X, Hagedorn CH, Cullen BR . Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs . RNA 2004 ; 10 : 1957 – 1966 .
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH et al . MicroRNA genes are transcribed by RNA polymerase II . EMBO J 2004 ; 23 : 4051 – 4060 .
Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC . Mammalian mirtron genes . Mol Cell 2007 ; 28 : 328 – 336 .
Pauley KM, Cha S, Chan EK . MicroRNA in autoimmunity and autoimmune diseases . J Autoimmun 2009 ; 32 : 189 – 194 .
Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC . The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila . Cell 2007 ; 130 : 89 – 100 .
Ruby JG, Jan CH, Bartel DP . Intronic microRNA precursors that bypass Drosha processing . Nature 2007 ; 448 : 83 – 86 .
Eulalio A, Huntzinger E, Izaurralde E . Getting to the root of miRNA-mediated gene silencing . Cell 2008 ; 132 : 9 – 14 .
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB . Prediction of mammalian microRNA targets . Cell 2003 ; 115 : 787 – 798 .
Lai EC . Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation . Nat Genet 2002 ; 30 : 363 – 364 .
Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y . Potent effect of target structure on microRNA function . Nat Struct Mol Biol 2007 ; 14 : 287 – 294 .
Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J et al . T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer . J Exp Med 2005 ; 201 : 1367 – 1373 .
Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K . Aberrant T cell differentiation in the absence of Dicer . J Exp Med 2005 ; 202 : 261 – 269 .
Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T et al . A role for Dicer in immune regulation . J Exp Med 2006 ; 203 : 2519 – 2527 .
Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C et al . Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage . Cell 2008 ; 132 : 860 – 874 .
O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ et al . A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway . Genes Dev 2007 ; 21 : 1999 – 2004 .
Chen CZ, Li L, Lodish HF, Bartel DP . MicroRNAs modulate hematopoietic lineage differentiation . Science 2004 ; 303 : 83 – 86 .
Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G et al . miR-181a is an intrinsic modulator of T cell sensitivity and selection . Cell 2007 ; 129 : 147 – 161 .
de Yébenes VG, Belver L, Pisano DG, González S, Villasante A, Croce C et al . miR-181b negatively regulates activation-induced cytidine deaminase in B cells . J Exp Med 2008 ; 205 : 2199 – 2206 .
Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J et al . MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb . Cell 2007 ; 131 : 146 – 159 .
Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF et al . Accumulation of miR-155 and BIC RNA in human B cell lymphomas . Proc Natl Acad Sci USA 2005 ; 102 : 3627 – 3632 .
Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N et al . Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice . Proc Natl Acad Sci USA 2006 ; 103 : 7024 – 7029 .
Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y et al . Regulation of the germinal center response by microRNA-155 . Science 2007 ; 316 : 604 – 608 .
Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K et al . Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein . Immunity 2009 ; 30 : 80 – 91 .
Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR et al . Requirement of bic/microRNA-155 for normal immune function . Science 2007 ; 316 : 608 – 611 .
Mattes J, Collison A, Plank M, Phipps S, Foster PS . Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease . Proc Natl Acad Sci USA 2009 ; 106 : 18704 – 18709 .
Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ et al . Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters . Cell 2008 ; 132 : 875 – 886 .
Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J et al . Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes . Nat Immunol 2008 ; 9 : 405 – 414 .
Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S et al . MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis . Nat Immunol 2009 ; 10 : 1252 – 1259 .
Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC . Ets-1 is a negative regulator of Th17 differentiation . J Exp Med 2007 ; 204 : 2825 – 2835 .
Otaegui D, Baranzini SE, Armañanzas R, Calvo B, Muñoz-Culla M, Khankhanian P et al . Differential micro RNA expression in PBMC from multiple sclerosis patients . PLoS One 2009 ; 4 : e6309 .
Furst DE, Keystone EC, Kirkham B, Kavanaugh A, Fleischmann R, Mease P et al . Updated consensus statement on biological agents for the treatment of rheumatic diseases, 2008 . Ann Rheum Dis 2008 ; 67 : iii2 – iii25 .
Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE et al . Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis . Arthritis Rheum 2008 ; 58 : 1001 – 1009 .
Abraham C, Cho JH . Inflammatory bowel disease . N Engl J Med 2009 ; 361 : 2066 – 2078 .
Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM et al . MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha . Gastroenterology 2008 ; 135 : 1624 – 1635 .
Wild S, Roglic G, Green A, Sicree R, King H . Global prevalence of diabetes: estimates for the year 2000 and projections for 2030 . Diabetes Care 2004 ; 27 : 1047 – 1053 .
Pandey AK, Agarwal P, Kaur K, Datta M . MicroRNAs in diabetes: tiny players in big disease . Cell Physiol Biochem 2009 ; 23 : 221 – 232 .
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE et al . A pancreatic islet-specific microRNA regulates insulin secretion . Nature 2004 ; 432 : 226 – 230 .
El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E . miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells . Diabetes 2008 ; 57 : 2708 – 2717 .
Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R . MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells . J Biol Chem 2006 ; 281 : 26932 – 26942 .
Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R et al . MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines . J Biol Chem 2007 ; 282 : 19575 – 19588 .
Acknowledgements
We thank Miss C. Liu for critically reading this manuscript. This work was supported by the Shanghai Natural Science Foundation (No. 10ZR1435300).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wei, B., Pei, G. microRNAs: critical regulators in Th17 cells and players in diseases. Cell Mol Immunol 7, 175–181 (2010). https://doi.org/10.1038/cmi.2010.19
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cmi.2010.19
Keywords
This article is cited by
-
MicroRNA-219a-5p suppresses intestinal inflammation through inhibiting Th1/Th17-mediated immune responses in inflammatory bowel disease
Mucosal Immunology (2020)
-
MicroRNA-125b regulates Th17/Treg cell differentiation and is associated with juvenile idiopathic arthritis
World Journal of Pediatrics (2020)
-
Upregulation of circulating microRNA-134 in adult-onset Still’s disease and its use as potential biomarker
Scientific Reports (2017)
-
Biomarkers and targeted new therapies for IgA nephropathy
Pediatric Nephrology (2017)
-
Inhibition of microRNA-155 ameliorates experimental autoimmune myocarditis by modulating Th17/Treg immune response
Journal of Molecular Medicine (2016)