Abstract
Pyruvate kinase M2 isoform (PKM2) catalyzes the last step of glycolysis and plays an important role in tumor cell proliferation. Recent studies have reported that PKM2 also regulates apoptosis. However, the mechanisms underlying such a role of PKM2 remain elusive. Here we show that PKM2 translocates to mitochondria under oxidative stress. In the mitochondria, PKM2 interacts with and phosphorylates Bcl2 at threonine (T) 69. This phosphorylation prevents the binding of Cul3-based E3 ligase to Bcl2 and subsequent degradation of Bcl2. A chaperone protein, HSP90α1, is required for this function of PKM2. HSP90α1's ATPase activity launches a conformational change of PKM2 and facilitates interaction between PKM2 and Bcl2. Replacement of wild-type Bcl2 with phosphorylation-deficient Bcl2 T69A mutant sensitizes glioma cells to oxidative stress-induced apoptosis and impairs brain tumor formation in an orthotopic xenograft model. Notably, a peptide that is composed of the amino acid residues from 389 to 405 of PKM2, through which PKM2 binds to Bcl2, disrupts PKM2-Bcl2 interaction, promotes Bcl2 degradation and impairs brain tumor growth. In addition, levels of Bcl2 T69 phosphorylation, conformation-altered PKM2 and Bcl2 protein correlate with one another in specimens of human glioblastoma patients. Moreover, levels of Bcl2 T69 phosphorylation and conformation-altered PKM2 correlate with both grades and prognosis of glioma malignancy. Our findings uncover a novel mechanism through which mitochondrial PKM2 phosphorylates Bcl2 and inhibits apoptosis directly, highlight the essential role of PKM2 in ROS adaptation of cancer cells, and implicate HSP90-PKM2-Bcl2 axis as a potential target for therapeutic intervention in glioblastoma.
Similar content being viewed by others
Introduction
Cancer cells exhibit increased level of reactive oxygen species (ROS) than normal cells partly due to oncogenic stimulation, increased metabolic activity and mitochondrial malfunction1,2,3. Under persistent oxidative stress, cancer cells become well-adapted to such stress through a set of mechanisms, which not only activate ROS-scavenging systems but also inhibit apoptosis4. A number of studies have suggested that such adaptation contributes to malignant transformation, metastasis and resistance to anticancer drugs5,6,7. Therefore, understanding mechanisms of ROS adaptation is important to efficiently kill cancer cells and overcome drug resistance.
Pyruvate kinase (PK) catalyzes the final step of glycolysis and is comprised of four isoenzymes encoded by two distinct genes in mammals, PKLR and PKM. Alternative splicing of PKM pre-mRNA results in the generation of pyruvate kinase M1 (PKM1) and M2 (PKM2) by the inclusion of exon 9 and exon 10, respectively8,9. PKM2, but not PKM1, is upregulated in most human cancers. Replacement of PKM2 with PKM1 in lung cancer cells inhibits aerobic glycolysis and tumor growth in nude mouse xenograft10. Activation of epidermal growth factor receptor (EGFR) in human glioma cells leads to increased glucose uptake and lactate production in a PKM2 expression-dependent manner11. Mediated by extracellular signal-regulated kinase (ERK)-dependent phosphorylation, PKM2 is capable of translocating to the nucleus upon EGRF activation12. In the nucleus, PKM2 binds to c-Src-phosphorylated β-catenin and enhances β-catenin's transactivation activity, promoting the expression of downstream oncogene cyclin D1 and the progression of cell cycle13. Under hypoxic conditions, prolyl-hydroxylated PKM2 interacts with HIF1a to induce glycolytic gene expression, which in turn enhances glucose metabolism in cancer cells14. These findings demonstrate the crucial roles of PKM2 in tumor cell proliferation.
Besides its important roles in promoting cell proliferation, PKM2 is also involved in the regulation of apoptosis. It has been shown that depletion of PKM2 expression by small interfering RNAs specifically against PKM2 results in decreased viability and increased apoptosis in multiple cancer cell lines15. Silencing of PKM2 in rat and human glioma spheroids enhances both apoptosis and differentiation16. In non-small cell lung cancer (NSCLC), PKM2 deficiency enhances ionizing radiation-induced apoptosis and autophagy in vitro and in vivo17. In addition to its anti-apoptotic function, PKM2 has been reported to translocate to the nucleus and induce a caspase-independent cell death in response to TT-23218. These results indicate that PKM2 also plays instrumental roles in cell survival. However, the mechanism underlying such roles of PKM2 remains elusive.
B-cell lymphoma 2 (Bcl2) is an anti-apoptotic member of the Bcl-2 family and is amplified in many human cancers19. The role of Bcl2 in ROS adaptation has been investigated in various laboratories. Ling et al.20 demonstrated that ROS generation plays a vital role in the initiation of apoptotic cascade by proteasome inhibitor bortezomib. Overexpression of Bcl2 in prostate carcinoma cells diminishes bortezomib-induced apoptosis. Hildeman et al.21 found that superoxide dismutase (SOD) mimetic MnTBAP decreases intracellular ROS and prevents apoptosis of activated T cells in vitro. Their study showed that MnTBAP increases the expression of Bcl2, which is normally decreased by T-cell activation21. These findings indicate that Bcl2 plays an important role in ROS adaptation in different cell types. However, the mechanism through which Bcl2 is regulated during such adaptation is not well understood.
In this study, we report that PKM2 translocates to the mitochondria under oxidative stress. Mitochondrial PKM2 interacts with and phosphorylates Bcl2, which then prevents Cul3-RBX1 ligase-mediated degradation of Bcl2, enhances apoptosis resistance of tumor cells and promotes gliomagenesis.
Results
PKM2 translocates to the mitochondria and inhibits hydrogen peroxide-induced Bcl2 degradation and apoptosis
To investigate PKM2's role in ROS adaptation, we depleted endogenous PKM2 in U87 or U251 human glioblastoma multiforme (GBM) cells through the infection of a lentivirus expressing a specific shRNA against PKM2 (Supplementary information, Figure S1A). After hydrogen peroxide (H2O2) or diamide (a thiol-oxidizing compound) treatment, U87 or U251 cells with PKM2 depletion (U87/shPKM2 or U251/shPKM2) had much more apoptotic cells than those cells expressing non-targeting shRNA (U87/shNT or U251/shNT), as determined by flow cytometry analysis of Annexin V-positive cells (Figure 1A and Supplementary information, Figure S1B). Similarly, caspase 3 activity was much more robust in U87/shPKM2 or U251/shPKM2 cells than that in U87/shNT or U251/shNT cells after H2O2 treatment (Figure 1B). Cytochrome c is released from the mitochondria to the cytosol, where it binds to Apaf1 to activate caspase cascades, during the early stage of mitochondria-dependent apoptosis22. Figure 1C showed that more cytochrome c was detected in cytosolic fraction in U87/shPKM2 or U251/shPKM2 cells than that in the cells expressing shNT after H2O2 treatment. Immune cells, such as Jurkat T cells, also express high levels of PKM2. To determine whether PKM2 has similar anti-apoptotic function in Jurkat T cells, we depleted endogenous PKM2 in Jurkat T cells and examined the effect of PKM2 depletion on H2O2-induced apoptosis. PKM2 depletion did not significantly increase the percentage of apoptotic cells in Jurkat T cells after H2O2 treatment (Supplementary information, Figure S1C), suggesting the cell-type specificity of PKM2 anti-apoptotic function. Taken together, these results demonstrate that PKM2 is essential for the resistance of glioma cells to oxidative stress-induced and mitochondria-dependent apoptosis.
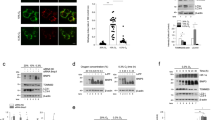
PKM2 translocates to the mitochondria and inhibits H2O2-induced Bcl2 degradation and apoptosis. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. Data are representative of at least three independent experiments. (A, B) U87 (top panel) or U251 (bottom panel) cells that stably express control shRNA (shNT) or shRNA against PKM2 (shPKM2) were treated with or without H2O2 (0.5 mM, 24 h). The cells were harvested for apoptosis analysis using Annexin V-Cy5 Apoptosis Kit (A) or caspase 3 activity analysis (B). Data represent the means ± SD of three independent experiments. CON, control. (C) U87 or U251 cells with or without PKM2 depletion were treated with H2O2 (0.5 mM, 24 h) and harvested for cytosolic (Cyto) and mitochondrial (Mito) fractionation. (D) U87 cells were treated with or without H2O2 (0.5 mM, 1 h). Immunofluorescence analyses were performed using anti-PKM2 or anti-VDAC antibodies. Representative images were shown in left panel. Co-localization of two proteins in 300 cells was quantified using the software of Volocity (PerkinElmer, right panel). (E) U87 or U251 cells were treated with or without H2O2 (0.5 mM, 1 h). Cytosolic and mitochondrial fractions of the cells were prepared. (F) U87/shNT or U87/shPKM2 cells were treated with or without H2O2 (0.5 mM, 4 h). (G) U87/shNT or U87/shPKM2 cells were treated with or without H2O2 (0.5 mM, 4 h). Quantitative RT-PCR was performed to examine mRNA levels of Bcl2. Data represent the means ± SD of three independent experiments. (H) U87/shPKM2 cells were pretreated with or without MG132 for 0.5 h, followed by treatment of H2O2 (0.5 mM) for 4 h. (I) U87/shNT or U87/shPKM2 cells were infected with a lentivirus expressing SFB-Bcl2. The cells were pretreated with MG132 for 0.5 h and then treated with or without H2O2 (0.5 mM, 4 h). Pull-down of SFB-Bcl2 was performed with streptavidin agarose beads. PD, pull-down.
To understand the mechanism of PKM2 regulation of such apoptosis, we examined subcellular localization of PKM2 after H2O2 treatment. Immunofluorescent analysis with anti-PKM2 or anti-VDAC (ubiquitously expressed and located in the outer mitochondrial membrane) antibody showed that ∼15% of PKM2 was translocated to the mitochondria in U87 cells upon H2O2 treatment (Figure 1D), which was supported by a subcellular fractionation analysis, showing that more PKM2 was detected in mitochondrial fraction of U87 or U251 cells after H2O2 treatment (Figure 1E). More thorough experiments of submitochondrial fractionation were also performed, showing that PKM2 was translocated to outer membrane of mitochondria upon H2O2 treatment (Supplementary information, Figure S1D). Thus, we speculated that PKM2 might regulate apoptosis via modulation of mitochondrial proteins. To test this possibility, we performed a mass spectrometry analysis of proteins associated with mitochondrial PKM2. Among them, three Bcl2 family proteins, including Bcl2, Bcl-xl and Bax, were identified (Supplementary information, Table S1). These proteins have been reported to regulate apoptosis by controlling mitochondrial membrane permeability19. Interactions between PKM2 and these proteins were further validated by co-immunoprecipitation analyses. Both Bcl2 and Bax could interact with PKM2, whereas Bcl-xl failed to do so (Supplementary information, Figure S1E). However, only Bcl2 protein level was markedly decreased in H2O2-treated U87/shPKM2 cells, whereas levels of Bcl-xl or Bax proteins were not (Figure 1F). mRNA levels of Bcl2 showed no significant difference between U87/shNT and U87/shPKM2 cells with or without H2O2 treatment, suggesting that PKM2 post-transcriptionally regulated Bcl2 (Figure 1G). Furthermore, treatment of a potent proteasome inhibitor MG132 abrogated H2O2-induced decrease of Bcl2 protein level in U87/shPKM2 cells (Figure 1H). These data confirmed that PKM2 depletion enhanced a proteasome-mediated degradation of Bcl2 induced by H2O2 treatment. Consistently, H2O2 induced more Bcl2 polyubiquitination in U87/shPKM2 cells than in U87/shNT cells (Figure 1I). These findings demonstrate that PKM2 translocates to the mitochondria and sustains Bcl2 protein stability, therefore enhancing the resistance of tumor cells to oxidative stress.
In addition to H2O2 treatment, we examined whether PKM2 depletion can enhance Bcl2 degradation under treatments of tumor necrosis factor α (TNFα; inflammatory cytokine), paclitaxel (PTX; anti-cancer cytotoxic drug) or temozolomide (TMZ; an alkylating agent used for glioma treatment). As shown in Supplementary information, Figure S1F, PKM2 depletion enhanced TNFα- and PTX-induced, but not TMZ-induced, Bcl2 degradation. More types of tumor cells, such as A549 lung cancer cells and HT29 colon cancer cells, were examined for PKM2's ability to stabilize Bcl2 protein. Consistent results were obtained, showing that PKM2 depletion promoted H2O2-induced degradation of Bcl2 in both A549 and HT29 cells (Supplementary information, Figure S1G).
To avoid the overexpression of target proteins, much less exogenous PKM2 or Bcl2 was expressed than those endogenous proteins in tumor cells through limiting infection efficiency (Supplementary information, Figure S1H).
PKM2 interacts with and stabilizes Bcl2
To investigate the mechanism through which PKM2 regulates Bcl2, a co-immunoprecipitation assay was performed to validate their interaction. Immunoblotting analyses of immunoprecipitated Bcl2 with an anti-PKM2 antibody (Figure 2A) or immunoprecipitated PKM2 with an anti-Bcl2 antibody (Supplementary information, Figure S2A) showed that endogenous PKM2 interacted with endogenous Bcl2 reciprocally, but not with succinate dehydrogenase (SDHA; used as a negative control) in both U87 and U251 cells upon H2O2 treatment. These observations were further supported by an immunofluorescence staining, showing that PKM2 colocalized with Bcl2 after H2O2 treatment (Figure 2B). We also examined the interaction between PKM2 and Bcl2 under various treatments or in various types of tumor cells. TNFα or PTX could induce the interaction between PKM2 and Bcl2, whereas TMZ failed to do so (Supplementary information, Figure S2B). Consistently, PKM2 interacted with Bcl2 in A549 and HT29 cells, but not in Jurkat T cells, upon H2O2 treatment (Supplementary information, Figure S2C). We further explored the location of PKM2 interaction with Bcl2, as PKM2 and Bcl2 also locate in the nucleus besides the mitochondria. Subcellular fractionation experiments were performed, showing that PKM2 interacted with Bcl2 in the mitochondria, but not in the nucleus (Supplementary information, Figure S2D).
PKM2 interacts with and stabilizes Bcl2. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. Data are representative of at least three independent experiments. (A) U87 or U251 cells were treated with or without H2O2 (0.5 mM, 1 h). Succinate dehydrogenase (SDHA) that has not been reported to interact with PKM2 or Bcl2 was used as a negative control. (B) U87 cells were treated with or without H2O2 (0.5 mM, 1 h). Immunofluorescence was performed using anti-PKM2 and anti-Bcl2 antibodies. Representative images were presented in left panel. Co-localization of two proteins in 300 cells was quantified using the software of Volocity (right panel). (C) U87 cells were infected with a lentivirus expressing Flag-PKM1 or Flag-PKM2 and then treated with or without H2O2 (0.5 mM, 1 h). (D) U87 cells were infected with a lentivirus expressing SFB-PKM2 WT or the indicated deletion mutants and then treated with or without H2O2 (0.5 mM, 1 h). Pull-down of SFB-PKM2 proteins was performed with streptavidin agarose beads. (E) U87 cells stably expressing SFB-PKM2 WT or ILLL were treated with or without H2O2 (0.5 mM, 1 h). Pull-down of SFB-PKM2 proteins was performed with streptavidin agarose beads. (F-H) U87 cells were depleted of endogenous PKM2 and then reconstituted with the expression of rPKM2 WT or rPKM2 ILLL (F). The cells were treated with or without H2O2 (0.5 mM) for 4 h (G, H). The cells were pretreated with MG132 for 0.5 h before H2O2 treatment (G). (I) U87 cells that stably express SFB-Bcl2 were treated with or without H2O2 (0.5 mM, 1 h). SFB-Bcl2 complex was precipitated using streptavidin agarose beads. A peptide competition assay was performed by mixing precipitated SFB-Bcl2 complex with 1.0 μg synthesized Flag or PKM2 389-405 peptides. (J, K) U87 cells that stably express SFB-Bcl2 were infected with a lentivirus expressing HA-tagged PKM2 389-405 peptide or Flag peptide and then treated with or without H2O2 (0.5 mM) for 1 (J) or 4 h (K). SFB-Bcl2 complex was precipitated using streptavidin agarose beads (J). Expression of Flag or PKM2 peptides was examined using dot blot assay with anti-HA antibody.
PKM1 shares high homology with PKM2 in protein sequence, as they differ from each other in only one exon. PKM2 contains exon 10, whereas PKM1 contains exon 99. To narrow searching range for Bcl2-interacting region on PKM2, we tested whether PKM1 interacts with Bcl2. U87 cells were infected with a lentivirus expressing Flag-tagged PKM2 (Flag-PKM2) or Flag-PKM1. Immunoblotting analyses of immunoprecipitated Flag-PKM2 or Flag-PKM1 with an anti-Bcl2 antibody showed that PKM1, unlike PKM2, did not interact with Bcl2 after H2O2 treatment (Figure 2C), suggesting that PKM2 may interact with Bcl2 through the region encoded by exon 10 of PKM2. According to secondary structure of PKM2 exon 10, we constructed four S-Flag-Streptavidin-binding peptide (SFB)-tagged PKM2 mutants with the deletion of the indicated amino acid residues, including PKM2 Δ370-388, PKM2 Δ389-405, PKM2 Δ406-423 and PKM2 Δ424-435. Immunoblotting analyses of precipitated SFB-PKM2 proteins with anti-Bcl2 antibody showed that only PKM2 Δ389-405 failed to bind to Bcl2 among PKM2 mutants (Figure 2D). Bcl2 has been reported to interact with other proteins through hydrophobic or ionic bonds23. According to amino acid properties, we constructed two PKM2 mutants, PKM2 ILLL, in which hydrophobic amino acids (isoleucine 389, leucine 392, leucine 394 and leucine 398) were mutated to alanine, and PKM2 EERR, in which ionic amino acids (glutamic acid 396, glutamic acid 397, arginine 399, and arginine 400) were mutated to alanine. Both PKM2 WT and PKM2 EERR interacted with Bcl2, whereas PKM2 ILLL failed to do so (Figure 2E and Supplementary information, Figure S2E).
To investigate whether PKM2-Bcl2 interaction is required for Bcl2 protein stability, we generated endogenous PKM2-depleted U87 cells with reconstituted expression of shRNA-resistant (r) PKM2 WT or rPKM2 ILLL (Figure 2F). Immunoblotting analyses of immunoprecipitated Bcl2 with anti-ubiquitin antibody showed that H2O2 treatment resulted in more polyubiquitination of Bcl2 in U87 cells expressing rPKM2 ILLL than in the cells expressing rPKM2 WT (Figure 2G). Consistently, Bcl2 protein levels were decreased in the cells expressing rPKM2 ILLL, but not in the cells expressing rPKM2 WT, after H2O2 treatment (Figure 2H).
To test the specificity of PKM2 ILLL mutation, we examined pyruvate kinase and protein kinase activities of PKM2 ILLL mutant. As shown in Supplementary information, Figure S2F, PKM2 ILLL has comparable pyruvate kinase activity to PKM2 WT. We further examined the glycolytic activities of the corresponding PKM2 ILLL-reconstituted cells. The data showed that similar glucose consumption and lactate production were detected in both PKM2 WT- and ILLL-reconstituted U87 cells (Supplementary information, Figure S2G). We previously reported that PKM2 could phosphorylate Histone H3 at threonine (T) 1124. Thus, we examined the phosphorylation of Histone H3 T11 by PKM2 WT or ILLL. The data showed that both PKM2 WT and ILLL could phosphorylate Histone H3 T11 at similar levels (Supplementary information, Figure S2H). Together, these data suggested that PKM2 ILLL only lost the binding to Bcl2, but still reserved its pyruvate kinase and protein kinase activities.
Peptide medicine is recognized for being highly selective and efficacious, and at the same time, relatively safe and well tolerated. We next investigated the possibility of the peptide that is composed of amino acids ranging from 389 to 405 of PKM2 as an anti-tumor drug. U87 cells were infected with a lentivirus expressing SFB-Bcl2. Precipitated SFB-Bcl2 complex was incubated with synthesized Flag or PKM2 389-405 peptides. PKM2 389-405 peptide remarkably disrupted H2O2-induced interaction between PKM2 and Bcl2, whereas Flag peptides could not (Figure 2I).
To confirm actual binding of PKM2 peptides to Bcl2, biotinylated PKM2 or Flag peptides were synthesized and incubated with U87 lysates or bacteria-purified recombinant Bcl2. As shown in Supplementary information, Figure S2I and S2J, endogenous Bcl2 or recombinant Bcl2 could be co-precipitated with biotinylated PKM2 peptides, but not with biotinylated Flag peptides, in a dose-dependent manner. In addition, the more PKM2 was expressed, the less Bcl2 was pulled down by biotinylated PKM2 peptides (Supplementary information, Figure S2K).
Inhibition of PKM2-Bcl2 interaction by PKM2 peptides was further examined in cells. U87 cells that stably express SFB-Bcl2 were infected with a lentivirus expressing PKM2 389-405 or Flag peptides. Expression of PKM2 peptides significantly inhibited H2O2-induced interaction between PKM2 and Bcl2, and caused subsequent degradation of Bcl2 (Figure 2J and 2K). Expression levels of PKM2 or Flag peptides were examined using dot blot assay, showing that PKM2 and Flag peptides had similar expression levels (Figure 2J and 2K). These results implicate a therapeutic potential of PKM2 389-405 peptide as an anti-cancer drug.
HSP90α1 alters PKM2 conformation and facilitates interaction between PKM2 and Bcl2
To investigate mechanism through which PKM2 interacts with Bcl2, we tested whether PKM2 interacts with Bcl2 directly. Recombinant His-Bcl2 and GST-PKM2 were purified from bacteria. Purities of two proteins were examined by gel staining with Coomassie Blue (Supplementary information, Figure S3A). GST pull-down assays were performed by mixing PKM2 and Bcl2 or PKM2 and commercial Histone H3 (NEB) that has been reported to interact with PKM2 in vitro24. The data showed that Bcl2 could not bind to PKM2 in vitro, whereas Histone H3 could do so (Figure 3A). Analysis of PKM2 structure implied that PKM2 389-405 fragment, especially four hydrophobic amino acids (I389, L392, L394 and L398) of PKM2 that are essential for PKM2-Bcl2 interaction, are mostly buried inside (Supplementary information, Figure S3B). Thus, we proposed that PKM2 structure should be altered to expose those critical amino acids. To search for potential isomerase, we performed a mass spectrum analysis of proteins associated with total PKM2 after H2O2 treatment. HSP90α1 was identified to be the only associated protein that can alter protein conformation (Supplementary information, Table S2). Interaction between HSP90α1 and PKM2 was further validated by a co-immunoprecipitation assay, showing that HSP90α1 interacted with PKM2 after H2O2 treatment (Figure 3B).
HSP90α1 alters PKM2 conformation and facilitates the interaction between PKM2 and Bcl2. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. Data are representative of at least three independent experiments. (A) GST pull-down assay was performed by mixing bacteria-purified recombinant GST-PKM2 and His-Bcl2, or GST-PKM2 and Histone H3 at 4 °C for 4 h. GST-PKM2 was precipitated using glutathione agarose beads. (B) U87 cells that stably express SFB-PKM2 were treated with or without H2O2 (0.5 mM, 1 h). SFB-PKM2 was pulled down using streptavidin agarose beads. (C) U87 cells that stably express shNT or shHSP90α1 were treated with or without H2O2 (0.5 mM, 1 h). (D) Flag-HSP90α1 WT and Flag-HSP90α1 deletion mutants, HSP90α1 ΔC(1-224), MD(1-544) and ΔN(545-731), used in the experiments are represented as black bars. (E) U87 cells were infected with a lentivirus expressing Flag-HSP90α1 WT or Flag-HSP90α1 deletion mutants and then treated with H2O2 (0.5 mM, 1 h). (F) U87 cells were infected with a lentivirus expressing Flag-HSP90α1 WT or Flag-HSP90α1 1-293 or Flag-HSP90α1 1-360 or Flag-HSP90α1 1-469 (bottom panel). Flag-HSP90α1 WT and Flag-HSP90α1 truncation mutants, HSP90α1 1-293, 1-360 and 1-469, used in the experiments are represented as black bars (top panel). (G) U87 cells were infected with a lentivirus expressing Flag-HSP90α1 WT or Flag-HSP90α1 Δ347-360. (H) Chaperone assay was performed by mixing GST-PKM2 and recombinant HSP90α1 WT or G97D. After the reaction, GST-PKM2 was pulled down using glutathione agarose beads. (I) U87 cells were depleted of endogenous HSP90α1 and then reconstituted with the expression of rHSP90α1 WT or G97D. The cells were further infected with a lentivirus expressing SFB-PKM2 and then treated with or without H2O2 (0.5 mM, 1 h). (J) HSP90α1-depleted U87 cells were reconstituted with the expression of rHSP90α1 WT or G97D and then treated with or without H2O2 (0.5 mM, 1 h). (K) Chaperone assay was performed by mixing recombinant HSP90α1 WT or G97D and GST-PKM2. After the reaction, GST-PKM2 were pulled down using glutathione agarose beads and incubated with His-Bcl2 at 4 °C for 4 h. (L) HSP90α1-depleted U87 cells that were reconstituted with the expression of rHSP90α1 WT or G97D were treated with or without H2O2 (0.5 mM, 4 h).
HSP90α1, an isoform of HSP90, is a chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress and aids in protein degradation. To determine whether HSP90α1 is required for PKM2-Bcl2 interaction, we depleted HSP90α1 in U87 cells with a shRNA against HSP90AA1 (Supplementary information, Figure S3C). Immunoblotting analyses of immunoprecipitated Bcl2 with anti-PKM2 antibody showed that HSP90α1 depletion abrogated the interaction between PKM2 and Bcl2 (Figure 3C). To map the interacting region of HSP90α1 and PKM2, three deletion mutants of HSP90α1 were constructed (Figure 3D), including N-terminal deletion mutant (ΔN), middle domain mutant (MD) and C-terminal deletion mutant (ΔC). Among three HSP90α1 mutants, only HSP90α1 MD could bind to PKM2 (Figure 3E). To further pinpoint the exact interacting region on HSP90α1, three more deletion mutants of HSP90α1 were constructed according to crystal structure of HSP90α125, including HSP90α1 1-293, 1-360 and 1-469. Co-immunoprecipitation analyses of HSP90α1 WT and mutants showed that only HSP90α1 1-293 lost the binding to PKM2 among the mutants under H2O2 treatment (Figure 3F). The region spanning amino acids 327-340 of HSP90β, which is homologous to the region spanning amino acids 347-360 of HSP90α1, has been reported to be involved in the binding to its client protein AKT26. Thus, we tested whether amino acids 347-360 of HSP90α1 are required for its binding to PKM2. HSP90α1 deletion mutant, HSP901 Δ347-360 lost the binding to PKM2 after H2O2 treatment (Figure 3G). Meanwhile, we also mapped the region of HSP90α1-PKM2 interaction on PKM2. Supplementary information, Figure S3D showed that both PKM2 Δ389-405 and PKM2 Δ406-423 lost their binding to HSP90α1. Ionic bonds are often involved in HSP90 substrate binding26. Considering simultaneous surface exposure of the interacting amino acids of PKM2, four ionic amino acids (R399, R400, D407 and E410) of PKM2, which are located on the surface of PKM2 protein, were mutated to alanine (PKM2 RRDE). Co-immunoprecipitation analyses showed that PKM2 WT interacted with HSP90α1 upon H2O2 treatment, whereas PKM2 RRDE mutant failed to do so (Supplementary information, Figure S3E). Together, these results demonstrated that amino acids 347-360 of HSP90α1 are involved in the binding to PKM2, and that PKM2 is likely a client protein of HSP90α1.
HSP90α1's ATPase activity often induces conformational change in client proteins, thereby causing their activation27. To determine whether HSP90α1 alters PKM2 conformation, we performed chaperon assay by mixing recombinant GST-PKM2 with HSP90α1 WT or HSP90α1 G97D (ATPase activity dead mutant), in which glycine 97 was mutated to aspartic acid. Immunoblotting analyses of precipitated GST-PKM2 with a PKM2-389 antibody, which specifically recognizes PKM2 389-405 fragment that is buried inside the protein before H2O2 treatment, showed that GST-PKM2 could only be recognized by PKM2-389 antibody in the presence of HSP90α1 WT, but not HSP90α1 G97D (Figure 3H), suggesting that PKM2 conformation was altered by HSP90α1 to expose PKM2 389-405 fragment. We further examined this result in cells. U87 cells were depleted of endogenous HSP90α1 and reconstituted with the expression of rHSP90α1 WT or rHSP90α1 G97D (Supplementary information, Figure S3F). Inhibition of HSP90 activity by HSP90α1 G97D expression was confirmed by examining the expression of HSP90α1 substrate proteins, AKT1 and Chk1. rHSP90α1 G97D expression significantly inhibited the expression of AKT1 and Chk1 (Supplementary information, Figure S3G). Immunoblotting analyses of the precipitated SFB-PKM2 with PKM2-389 antibody showed that H2O2-induced alteration of PKM2 conformation could only be detected in the cells expressing rHSP90α1 WT, but not in the cells expressing rHSP90α1 G97D (Figure 3I).
To determine whether HSP90α1-mediated conformational change of PKM2 is required for PKM2-Bcl2 interaction, co-immunoprecipitation assay was performed using Bcl2 antibody in HSP90α1-depleted U87 cells that were reconstituted with the expression of rHSP90α1 WT or G97D. As shown in Figure 3J, PKM2 interacted with Bcl2 in cells expressing rHSP90α1 WT, but not in cells expressing rHSP90α1 G97D, after H2O2 treatment. This result was further supported by an HSP90α1-mediated chaperon assay followed by a GST pull-down assay, showing that PKM2 could only interact with Bcl2 in the presence of HSP90α1 WT, but not HSP90α1 G97D (Figure 3K). Consistently, H2O2 treatment induced more degradation of Bcl2 in the cells expressing rHSP90α1 G97D than in the cells expressing rHSP90α1 WT (Figure 3L).
HSP90α1-mediated conformational change of PKM2 was also confirmed using an HSP90 inhibitor (17-AAG), which could efficiently inhibit expression of AKT1 and Chk1 (Supplementary information, Figure S3H). As shown in Supplementary information, Figure S3I and S3J, pretreatment of 17-AAG markedly inhibited H2O2-induced alteration of PKM2 conformation and PKM2-Bcl2 interaction. Consistently, H2O2-induced degradation of Bcl2 was markedly enhanced by 17-AAG treatment (Supplementary information, Figure S3K).
PKM2 phosphorylates Bcl2 at T69
It has been reported that Bcl2 stability could be regulated by phosphorylation28. To investigate whether PKM2 regulates Bcl2 phosphorylation, we performed mass spectrometry analysis of Bcl2 phosphorylation in U87 cells after H2O2 treatment, showing that threonine (T) 69 and serine (S) 70 of Bcl2 were phosphorylated after H2O2 treatment. Immunoblotting analyses of the precipitated SFB-Bcl2 with anti-Bcl2 pT69 and anti-Bcl2 pS70 antibodies showed that PKM2 depletion significantly blocked Bcl2 T69 phosphorylation, but not Bcl2 S70 phosphorylation (Figure 4A and Supplementary information, Figure S4A). Regulation of Bcl2 T69 phosphorylation by PKM2 was further verified in more types of tumor cells or under more treatments. Both TNFα- and PTX-induced Bcl2 T69 phosphorylation were significantly inhibited by PKM2 depletion, whereas TMZ treatment could not even induce Bcl2 T69 phosphorylation (Supplementary information, Figure S4B). H2O2-induced Bcl2 T69 phosphorylation was blocked by PKM2 depletion in A549 or HT29 cells, whereas Bcl2 T69 phosphorylation could not even be induced in Jurkat T cells after H2O2 treatment (Supplementary information, Figure S4C).
PKM2 phosphorylates Bcl2 at T69. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. Data are representative of at least three independent experiments. (A) U87 cells that stably express shNT or shPKM2 were infected with a lentivirus expressing SFB-Bcl2 and then treated with or without H2O2 (0.5 mM, 1 h). SFB-Bcl2 was pulled down using streptavidin agarose beads. (B) Chaperon assay was performed with recombinant HSP90α1 and GST-PKM2. After reaction, GST-PKM2 was pulled down using glutathione agarose beads. In vitro kinase assays were performed by mixing isomerized GST-PKM2 and His-Bcl2 WT or Bcl2 T69A or Bcl2 S70A. (C) Chaperon assay was performed with recombinant HSP90α1 and GST-PKM2 or GST-PKM1. GST-PKM2 or GST-PKM1 was pulled down using glutathione agarose beads. In vitro kinase assay was performed by mixing His-Bcl2 and isomerized GST-PKM2 or GST-PKM1. (D) Chaperon assay was performed with recombinant HSP90α1 and GST-PKM2. GST-PKM2 was pulled down using glutathione agarose beads. In vitro kinase assays were performed by mixing isomerized GST-PKM2 and His-Bcl2. After reaction, His-Bcl2 was pulled down using Nickel agarose beads and analyzed by mass spectrometry. Mass spectrometric analysis of a tryptic fragment at m/z 182.054 (mass error was 0.10 p.p.m.) matched to the doubly charged peptide 69-pho-tSPLQTPAAPGAAAGPALSPVPPVVHLTLR-98, suggesting that T69 was phosphorylated. The Sequest score for this match was Xcorr = 2.18; Mascot scores were 27, and expectation value was 5.1 × 10−4. The Best score evidence ID was 460, and site probability was 78.4%. (E) PKM2-depleted U87 cells that were reconstituted with the expression of rPKM2 WT or ILLL were infected with a lentivirus expressing SFB-Bcl2 and then treated with or without H2O2 (0.5 mM, 1 h). (F, G) U87 cells that stably express SFB-Bcl2 WT or T69A were treated with or without H2O2 (0.5 mM, 4 h). The cells were pretreated with MG132 for 0.5 h before H2O2 treatment (F).
Previous studies from our and other groups have reported that PKM2 is capable of phosphorylating proteins as a protein kinase24. We next tested whether PKM2 phosphorylates Bcl2. In vitro kinase assays were performed by mixing 32p-labeled PEP with recombinant PKM2 or PKM1 and Bcl2 WT or phosphorylation-deficient Bcl2 mutants. Figure 4B and 4C showed that PKM2, but not PKM1, could phosphorylate Bcl2, and that Bcl2 T69A was resistant to phosphorylation, whereas Bcl2 S70A could still be phosphorylated. Phosphorylation of Bcl2 T69 by PKM2 was further validated by LC-MS/MS analyses, showing that Bcl2 T69 was the only residue phosphorylated by PKM2 (Figure 4D and Supplementary information, Figure S4D). To further verify Bcl2 T69 phosphorylation by PKM2 in cells, U87 cells with depleted PKM2 and reconstituted expression of rPKM2 WT or ILLL were infected with a lentivirus expressing SFB-Bcl2. Immunoblotting analyses of the precipitated SFB-Bcl2 with anti-Bcl2 pT69 antibody showed that Bcl2 T69 was phosphorylated in cells expressing rPKM2 WT, but not in cells expressing rPKM2 ILLL, after H2O2 treatment (Figure 4E). Together, these data clearly indicated that PKM2 phosphorylated Bcl2 at T69 after H2O2 treatment.
We further tested whether PKM2-dependent Bcl2 phosphorylation is required to sustain Bcl2 protein level. U87 cells were infected with a lentivirus expressing SFB-Bcl2 WT or T69A. Immunoblotting analyses of the precipitated SFB-Bcl2 with an anti-ubiquitin antibody showed that Bcl2 T69A had more polyubiquitination than Bcl2 WT after H2O2 treatment (Figure 4F). Consistently, H2O2 treatment resulted in more degradation of Bcl2 T69A than that of Bcl2 WT (Figure 4G).
PKM2-dependent Bcl2 T69 phosphorylation prevents the binding of Cul3-based E3 ligase to Bcl2
It has been reported that Bcl2 could be ubiquitinated by Cul3-based E3 ligase and subsequently degraded29. To investigate the mechanism through which PKM2 stabilizes Bcl2 protein, we examined whether PKM2 interferes with recruitment of Cul3-based E3 ligase to Bcl2. U87 cells were depleted of PKM2 and reconstituted with expression of rPKM2 WT or ILLL. Immunoblotting analyses of immunoprecipitated Bcl2 with anti-Cul3 and anti-Rbx1 antibodies showed that Bcl2 interacted with both Cul3 and Rbx1 in cells expressing rPKM2 ILLL, but not in cells expressing rPKM2 WT, after H2O2 treatment (Figure 5A), suggesting that PKM2 interacts with Bcl2 and prevents binding of E3 ligase to Bcl2. We next examined whether Bcl2 T69 phosphorylation is required to prevent the recruitment of E3 ligase. Figure 5B showed that Bcl2 T69A was associated with more Cul3 and Rbx1 than Bcl2 WT after H2O2 treatment. In contrast, phosphorylation-mimic mutant of Bcl2, Bcl2 T69E, interacted with much less Cul3 and Rbx1 than Bcl2 WT in H2O2-treated U87 cells with PKM2 depletion (Figure 5C).
PKM2-dependent Bcl2 T69 phosphorylation prevents the binding of Cul3-based E3 ligase to Bcl2. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. Data are representative of at least three independent experiments. (A) PKM2-depleted U87 cells were reconstituted with the expression of rPKM2 WT or ILLL and then treated with or without H2O2 (0.5 mM, 1 h). (B) U87 cells that stably express SFB-Bcl2 WT or T69A were treated with or without H2O2 (0.5 mM, 1 h). SFB-Bcl2 was pulled down using streptavidin agarose beads. (C) PKM2-depleted U87 cells were infected with a lentivirus expressing SFB-Bcl2 WT or T69E and then treated with or without H2O2 (0.5 mM, 1 h). SFB-Bcl2 was pulled down using streptavidin agarose beads. (D, F) PKM2-depleted U87 cells were reconstituted with the expression of rPKM2 ILLL and then transfected with or without siRNA against CUL3 (siCul3). The cells were treated with or without H2O2 (0.5 mM, 4 h). The cells were pretreated with MG132 for 0.5 h (D). (E, G) U87 cells that stably express SFB-Bcl2 T69A were transfected with or without siCul3 and then treated with or without H2O2 (0.5 mM, 4 h). The cells were pretreated with MG132 before H2O2 treatment (E).
To further demonstrate whether Cul3-based E3 ligase is involved in H2O2-induced Bcl2 degradation, we depleted Cul3 using siRNA against CUL3 (siCul3) in U87 cells with depleted PKM2 and reconstituted expression of rPKM2 ILLL, or in U87 cells expressing Bcl2 T69A. Immunoblotting analyses of immunoprecipitated Bcl2 or precipitated SFB-Bcl2 with anti-ubiquitin antibodies showed that Cul3 depletion remarkably inhibited H2O2-induced polyubiquitination of Bcl2 in cells expressing rPKM2 ILLL or H2O2-induced polyubiquitination of Bcl2 T69A (Figure 5D and 5E). Consistently, Cul3 depletion blocked H2O2-induced degradation of Bcl2 in cells expressing rPKM2 ILLL or H2O2-induced degradation of Bcl2 T69A (Figure 5F and 5G). Taken together, these results demonstrate that PKM2-dependent phosphorylation of Bcl2 T69 prevents the binding of Cul3-Rbx1 ligase to Bcl2, therefore inhibiting Bcl2 degradation.
Phosphorylation of Bcl2 T69 by PKM2 enhances resistance of tumor cells to oxidative stress and promotes gliomagenesis
To explore the biological function of PKM2-dependent Bcl2 T69 phosphorylation, U87 or U251 cells were depleted of PKM2 and reconstituted with the expression of rPKM2 WT or ILLL (Figure 2F and Supplementary information, Figure S5A); U87 or U251 cells were depleted of Bcl2 and reconstituted with the expression of rBcl2 WT or T69A (Supplementary information, Figure S5B and S5C). Apoptosis analysis using Annexin V-Cy5 staining showed that more apoptotic cells were detected in cells expressing rPKM2 ILLL or rBcl2 T69A than in cells expressing rPKM2 WT or rBcl2 WT after H2O2 treatment (Figure 6A and 6B, Supplementary information, Figure S5D and S5E, top panel). Consistently, H2O2 treatment led to the stronger caspase 3 activity in cells expressing rPKM2 ILLL or rBcl2 T69A than that in cells expressing rPKM2 WT or rBcl2 WT after H2O2 treatment (Figure 6A and 6B, Supplementary information, Figure S5D and S5E, bottom panel). In addition, effects of phosphorylation-mimic Bcl2 T69E on apoptosis were examined. As shown in Supplementary information, Figure S5F, expression of rBcl2 T69E markedly abrogated H2O2-induced apoptosis of PKM2-depleted U87 cells. Together, these data demonstrate that PKM2-dependent Bcl2 T69 phosphorylation is essential for oxidative stress adaptation of glioma cells.
Phosphorylation of Bcl2 T69 by PKM2 enhances the resistance of tumor cells to oxidative stress and promotes gliomagenesis. Immunoblotting analyses were performed with the indicated antibodies. Data are representative of at least three independent experiments. (A) U87 cells were depleted of endogenous PKM2 and then reconstituted with the expression of rPKM2 WT or ILLL. The cells were treated with or without H2O2 (0.5 mM, 24 h). The cells were harvested for analyses of apoptosis using Annexin V staining (top panel) or caspase 3 activity (bottom panel). Data represent the means ± SD of three independent experiments. (B) U87 cells were depleted of endogenous Bcl2 and then reconstituted with the expression of rBcl2 WT or T69A. The cells were treated with or without H2O2 (0.5 mM, 24 h). The cells were harvested for analyses of apoptosis using Annexin V staining (top panel) or caspase 3 activity (bottom panel). Data represent the means ± SD of three independent experiments. (C, D) U87 cells were depleted of endogenous Bcl2 and reconstituted with the expression of rBcl2 WT or T69A (C). U87 cells were depleted of endogenous PKM2 and reconstituted with expression of rPKM2 WT or ILLL (D). Endogenous PKM2-depleted U87 cells with reconstituted expression of rPKM2 ILLL were further infected with the lentivirus expressing Flag-Bcl2 WT or T69E (D). All genetically-modified U87 cells above were harvested and intracranially injected into randomized athymic nude mice (seven mice per group). After 40 days, the mice were euthanized and tumor growth was examined. H&E-stained coronal brain sections show representative tumor xenografts (top panel). Representative images of tumor boundaries were presented in middle panel with 200× magnification. Tumor volumes were measured using length (a) and width (b) and calculated using the equation: V = ab2/2 (bottom panel). Data represent the means ± SD of seven mice. (E) Kaplan-Meier survival curves for the mice intracranially injected with Bcl2-depleted U87 cells reconstitutively expressing rBcl2 WT or T69A, or PKM2-depleted U87 cells reconstitutively expressing rPKM2 WT or ILLL, or Bcl2 WT- or T69E-expressing U87 cells that were depleted of PKM2 and reconstituted with the expression of rPKM2 ILLL (10 mice per group). (F, G) Tumor tissues derived from the mice injected with PKM2-depleted U87 cells reconstitutively expressing rPKM2 WT or ILLL (F), or Bcl2-depleted U87 cells reconstitutively expressing rBcl2 WT or T69A (G) were dissected for immunoblotting analyses. (H, J) U87 cells were intracranially injected into randomized athymic nude mice. After 15 days, 7 mice were sacrificed to examine tumor growth; the remaining mice were intratumorally injected with the lentivirus expressing Flag or PKM2 (389-405) peptides, and then sacrificed on day 40. H&E-stained coronal brain sections show representative tumor xenografts (H, top panel). Representative images of tumor boundaries were presented in middle panel with 200× magnification. Tumor volumes were calculated (H, bottom panel). Data represent the means ± SD of 7 mice. Tumor tissues derived from the mice injected with the lentivirus expressing Flag or PKM2 389-405 peptides were dissected for immunoblotting analyses (J). (I) U87 cells were intracranially injected into randomized athymic nude mice. After 15 days, the mice were intracranially injected with the lentivirus expressing Flag or PKM2 (389-405) peptides. Kaplan-Meier survival curves were then plotted.
To further explore the cellular functions of PKM2-dependent Bcl2 phosphorylation, apoptosis analyses were performed in U87 cells under various types of treatments. As shown in Supplementary information, Figure S5G and S5H, more apoptotic cells were observed in PKM2-depleted U87 cells with reconstituted expression of rPKM2 ILLL or Bcl2-depleted U87 cells with reconstituted expression of rBcl2 T69A than those in the cells expressing rPKM2 WT or rBcl2 WT after TNFα or PTX treatment. It has been extensively reported that the treatment of TNFα or PTX induces significant ROS production and ROS plays an essential role in TNFα- or PTX-induced apoptosis30,31. Thus, PKM2-dependent Bcl2 regulation induced by TNFα or PTX is very likely mediated by increased ROS after the treatment. This hypothesis was simply tested by using the antioxidant N-acetyl-cysteine (NAC) to decrease intracellular ROS levels. The data showed that NAC treatment significantly abrogated TNFα- or PTX-induced apoptosis in PKM2-depleted U87 cells, and that TNFα- or PTX-induced Bcl2 phosphorylation at T69 was also inhibited by NAC treatment (Supplementary information, Figure S5I and S5J). These results indicate that TNFα- or PTX-induced Bcl2 regulation by PKM2 was ROS dependent, suggesting a specific role of PKM2 under oxidative stress.
PKM2-dependent Bcl2 phosphorylation was also investigated in other types of tumor cells. A549 or HT29 cells were depleted of endogenous PKM2 and reconstituted with rPKM2 WT or ILLL, or were depleted of endogenous Bcl2 and reconstituted with rBcl2 WT or T69A. Consistently, H2O2 treatment induced more apoptosis in A549 or HT29 cells expressing rPKM2 ILLL or rBcl2 T69A than in the cells expressing rPKM2 WT or rBcl2 WT (Supplementary information, Figure S5K-S5N).
Conditions associated with tumorigenesis (including gliomagenesis) such as hypoxia, matrix detachment, mitochondrial dysfunction and inflammation can all lead to excess production of ROS32,33,34. Therefore, cancer cells are particularly challenged while dealing with oxidative stress35,36. To evaluate the contribution of PKM2-dependent Bcl2 T69 phosphorylation to brain tumor development, we intracranially injected (1) Bcl2-depleted U87 cells with reconstituted expression of rBcl2 WT or T69A, (2) PKM2-depleted U87 cells with reconstituted expression of rPKM2 WT or ILLL, or (3) Bcl2 WT- or T69E-expressing U87 cells that were depleted of PKM2 and reconstituted with the expression of rPKM2 ILLL (Supplementary information, Figure S5O) into randomized athymic nude mice. Analyses of hematoxylin and eosin (H&E) staining and tumor volume showed that cells expressing rPKM2 WT or rBcl2 WT elicited rapid tumorigenesis. In contrast, brain tumor development was markedly dampened by the expression of rPKM2 ILLL or rBcl2 T69A. Notably, rPKM2 ILLL expression-suppressed tumorigenesis was restored by Bcl2 T69E expression, but not Bcl2 WT expression (Figure 6C and 6D). Clear boundaries between tumor cells and normal cells without obvious inflammatory lesions were observed in the brain sections from the mice inoculated with genetically-modified U87 cells (Figure 6C and 6D, middle panel). Kaplan-Meier survival curves showed that mice injected with the cells expressing rBcl2 T69A or rPKM2 ILLL survived longer time than mice injected with the cells expressing rBcl2 WT or rPKM2 WT. Consistently, the expression of Bcl2 T69E, but not Bcl2 WT, could significantly shorten the survival time of the mice inoculated with PKM2 ILLL-reconstituted U87 cells (Figure 6E).
Further, the contribution of PKM2-dependent Bcl2 T69 phosphorylation to gliomagenesis was evaluated using luciferase-labeled U87 cells. Quantification of real-time imaging of tumor growth showed that PKM2-depleted U87 cells expressing rPKM2 WT or Bcl2-depleted cells expressing rBcl2 WT expanded quickly in mice 15 days after implantation and reached maximum number after 40 days, whereas cells expressing rPKM2 ILLL or rBcl2 T69A failed to do so (Supplementary information, Figure S5P and S5Q). Consistent results were also obtained in mice implanted with (1) PKM2-depleted U251 cells with reconstituted expression of rPKM2 WT or rPKM2 ILLL, and (2) Bcl2-depleted U251 cells with reconstituted expression of rBcl2 WT or T69A. Kaplan-Meier survival curves showed that mice injected with U251 cells expressing rPKM2 ILLL or rBcl2 T69A had longer survival time than mice injected with U251 cells expressing rPKM2 WT or rBcl2 WT (Supplementary information, Figure S5R). Furthermore, immunoblotting analyses of the dissected tumor tissues showed that decreased levels of Bcl2 and increased levels of cleaved caspase 3 were detected in tumor tissues derived from the mice injected with U87 cells expressing rPKM2 ILLL or rBcl2 T69A compared to those in the counterpart tissue derived from the mice injected with U87 cells expressing rPKM2 WT or rBcl2 WT (Figure 6F and 6G). Together, these results demonstrate that Bcl2 T69 phosphorylation by PKM2 is crucial for brain tumor development.
We next investigated therapeutic effects of PKM2 389-405 peptide on gliomagenesis. U87 cells were intracranially injected into randomized nude mice. After 15 days, the mice bearing relatively small tumors were intratumorally injected with a lentivirus expressing Flag or PKM2 389-405 peptides. On day 40, analyses of H&E staining and tumor volume showed that expression of PKM2 389-405 peptide, but not Flag peptide, significantly impaired tumor growth (Figure 6H). Kaplan-Meier survival curves showed the mice injected with the lentivirus expressing PKM2 389-405 peptide had much longer survival time than the mice injected with the lentivirus expressing Flag peptides (Figure 6I). Luciferase-labeled U87 cells were also used to evaluate therapeutic potential of PKM2 peptides. As shown in Supplementary information, Figure S5S, luciferase-expressing U87 cells grew into a small tumor 15 days after implantation. On day 40, the mice injected with the lentivirus expressing PKM2 389-405 peptide bear much smaller tumor than the mice injected with the lentivirus expressing Flag peptides. Consistent results were obtained in mice implanted with U251 cells followed by the injection of the lentivirus expressing Flag peptide or PKM2 389-405 peptide, showing that the expression of PKM2 peptide markedly prolonged mouse survival (Supplementary information, Figure S5T). Immunoblotting analyses of the dissected tumor tissues showed decreased level of Bcl2 and increased level of cleaved caspase 3 in tumor tissues derived from the mice injected with the lentivirus expressing PKM2 389-405 peptide compared to those in the counterpart tissues derived from the mice injected with the lentivirus expressing Flag peptides (Figure 6J). Together, these results demonstrate that PKM2 389-405 peptide could inhibit gliomagenesis effectively, highlighting therapeutic potential of PKM2 peptide as an anti-tumor drug.
Bcl2 pT69 levels positively correlate with levels of conformation-altered PKM2 and grades of glioma malignancy and prognosis
To investigate clinical significance of PKM2-dependent Bcl2 T69 phosphorylation, immunohistochemistry (IHC) analyses were performed with 40 specimens from human primary GBM patients (World Health Organization (WHO) grade IV) using antibodies of anti-Bcl2, anti-Bcl2 pT69 or anti-PKM2-389, which specifically recognizes PKM2 with conformational change. Specificities of the antibodies used for IHC staining were validated by a blocking peptide/recombinant protein competition assay (Supplementary information, Figure S6). As shown in Figure 7A, Bcl2 protein levels correlated with levels of conformation-altered PKM2 or Bcl2 pT69. Pearson correlation analysis of the staining on a scale of 0-8.0 showed that these correlations were significant (Figure 7B and 7C).
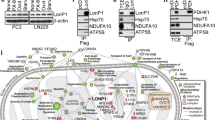
Bcl2 pT69 levels positively correlates with levels of conformation-altered PKM2 and grades of glioma malignancy and prognosis. (A-C) Immunohistochemical analyses of 40 specimens from GBM patients using anti-PKM2-389, anti-Bcl2 pT69 and anti-Bcl2 antibodies were performed. Representative images of two GBM specimens (A). Semi-quantitative scoring (using a scale from 0 to 8) was carried out (Pearson product moment correlation test; B, r = 0.76, P < 0.001; C, r = 0.81, P < 0.001). (D, E) Survival of 80 patients with low (0-4.5 staining scores, blue curve) versus high (4.6-8 staining scores, red curve) conformation-altered PKM2 levels (low, 22 patients; high, 58 patients) was analyzed (D). Survival of 80 patients with low (0-3.9 staining scores, blue curve) versus high (4.0-8 staining scores, red curve) Bcl2 T69 phosphorylation levels (low, 28 patients; high, 52 patients) was compared (E). Empty circles represent the deceased patients, and filled circles represent the censored (alive at last clinical follow-up) patients. (F, G) Forty diffuse astrocytoma specimens were immunohistochemically stained using PKM2-389 antibody (F) or anti-Bcl2 pT69 antibody (G). Staining scores of the specimens were compared with 40 (F) or 80 (G) stained GBM specimens (Student's t-test, two tailed, P < 0.001). (H) Schematic representation of the PKM2-mediated Bcl2 stabilization and apoptosis resistance. PKM2 translocates to the mitochondria under oxidative stress, where PKM2 interacts with and phosphorylates Bcl2 at T69. Bcl2 T69 phosphorylation that requires HSP90α1-mediated PKM2 isomerization prevents the binding of Cul3-based E3 ligase to Bcl2, which stabilizes Bcl2 and therefore inhibits apoptosis and promotes tumorigenesis.
We next examined the correlation of survival of 80 GBM patients, all of whom received standard adjuvant radiotherapy after surgery and the following treatment with an alkylating agent (TMZ in most cases), with levels of Bcl2 pT69 (low, 0-4.5 staining score; high, 4.6-8 staining score) or levels of PKM2 with conformational change (low, 0-3.9 staining score; high, 4.6-8 staining score). Patients whose tumors had low levels of conformation-altered PKM2 (22 cases) had a median survival of 169.58 weeks, whereas those whose tumors had high levels of conformation-altered PKM2 (58 cases) had a significantly lower median survival of 69.33 weeks (Figure 7D). Patients whose tumors had low levels of Bcl2 pT69 (28 cases) had a median survival of 166.66 weeks, whereas those whose tumors had high levels of Bcl2 pT69 (52 cases) had a significantly lower median survival of 59.34 weeks (Figure 7E). Correlations of levels of Bcl2 pT69 and conformation-altered PKM2 with grades of glioma malignancy were also explored. Levels of conformation-altered PKM2 or Bcl2 pT69 in samples from patients with low-grade diffuse astrocytoma (WHO grade II; median survival time > 5 years) were compared with those from patients with high-grade GBM37. IHC analyses showed that levels of conformation-altered PKM2 or Bcl2 pT69 in low-grade tumors were significantly lower than those in GBM tumors (Figure 7F and 7G).
Taken together, these results clarify the importance of PKM2-dependent Bcl2 phosphorylation and stabilization in the clinical behavior of human GBM and reveal a relationship between Bcl2 T69 phosphorylation or conformation alteration of PKM2 and clinical glioma malignancy.
Discussion
PKM2 is upregulated in many human cancers and promotes tumor cell proliferation through multiple mechanisms. Recent studies reported that PKM2 also regulates apoptosis. However, mechanisms underlying such a role of PKM2 remains unknown. Here we domonstrate that oxidative stress induces mitochondrial translocation of PKM2. In the mitochondria, PKM2, functioning as a protein kinase, interacts with and phosphorylates Bcl2 at T69, which disrupts assocation of Cul3-based E3 ligase with Bcl2, and blocks proteasome-mediated Bcl2 degradation, thereby inhibiting oxidative stress-induced apoptosis (Figure 7H). These results reveal a novel mechanism by which PKM2 regulates apoptosis directly, and highlight the critical role of PKM2 in ROS adaptation of tumor cells. Importantly, levels of Bcl2 T69 phosphorylation and conformation-altered PKM2 correlate with grades of glioma malignancy and prognosis of GBM patient, suggesting that Bcl2 T69 phosphorylation and conformational change of PKM2 could be two biomarkers to predict brain tumor malignancy and prognosis. Furthermore, the finding that PKM2 389-405 peptide disrupts PKM2-Bcl2 interaction, promotes Bcl2 degradation, and hampers brain tumor development points to the potential of this peptide as an anti-cancer drug.
Elevated levels of ROS have been detected in almost all cancer types. Cancer cells can adapt to such stress through multiple mechanisms38. Here we demonstrate that PKM2 is involved in such adaptation by inhibiting apoptosis. Our data show that increased level of ROS induced mitochondrial translocation of PKM2. Mitochondrial PKM2 interacts with and phosphorylates Bcl2. PKM2-dependent Bcl2 phosphorylation prevents Bcl2 degradation and thereby inhibits ROS-induced apoptosis. In addition to apoptosis inhibition, cancer cells also activate ROS-scavenging systems to cope with increased ROS levels4. PKM2 is also involved in this process. Increased intracellular ROS level causes inhibition of glycolytic activity of PKM2 through oxidation of cysteine 358, which diverts glucose flux into the pentose phosphate pathway, thereby generating sufficient reducing potential for detoxification of ROS39. Thus, PKM2 can promote the adaptation of cancer cells to ROS at two levels, suggesting therapeutic potential of targeting PKM2.
PKM2 can be regulated at multiple levels, such as posttranslational modification and subcellular localization. It has been shown in our previous studies that PKM2 is phospohorylated at serine 37 by ERK 1 and 2 upon EGFR activation12. PKM2 S37 phosphorylation promotes nuclear translocation of PKM2. In the nucleus, PKM2 interacts with β-catenin and thereby enhances transactivation activity of β-catenin, which enhances downstream oncogene expression13. Nuclear translocation of PKM2 was also observed by Lv et al.40, showing that PKM2 K433 acetylation that was mediated by the p300 acetyltransferase prevents the binding of FBP to PKM2 and thereby facilitates nuclear translocation of PKM2 and its switch from a cytoplasmic metabolic enzyme to a nuclear protein kinase. Both studies demonstrated nuclear localization and functions of PKM2. In this study, we showed for the first time that PKM2 translocates to the mitochondria under oxidative stress. Submitochondrial fractionation analyses showed that PKM2 translocated to outermembrane of mitochondria (Supplementary information, Figure S1D). It is likely that PKM2-Bcl2 interaction bridges PKM2 to mitochondrial outermembrane, which remains to be clarified.
Protein kinase activity of PKM2 has been extensively reported. PKM2 phosphorylates Histone H3 or STAT3 in the nucleus and regulates various gene expression24,41. Bub3 and myosin light chain 2 are also phosphorylated by PKM2 to promote mitosis and cytokinesis in the context of cell cycle42,43. Moreover, protein kinase activity of PKM2 on Histone H3 can be enhanced by succinyl-5-aminoimidazole-4-carboxamide-1-ribose-5′-phosphate (SAICAR) and over 100 proteins were identified to be phosphorylated by PKM244. Notably, protein kinase activity of PKM2 was further supported by two recent publications, showing (1) that yeast PKM2 homolog directly phosphorylates histone H3 at T11 both in vivo and in vitro, and (2) that nuclear PKM2 phosphorylates Stat3 and promotes proinflammatory phenotype of macrophages in coronary artery disease through boosting IL-6 and IL-1β production45,46. Together, these studies demonstrate the protein kinase activity of PKM2 in the nucleus. Here we showed that oxidative stress stimulates the protein kinase activity of PKM2 to phosphosrylate Bcl2 at T69 in the mitochondria, which is essential for the anti-apoptotic function of PKM2.
Roles of Bcl2 can be regulated by phosphorylation. SLAMF1 deficiency in chronic lymphocytic leukemia (CLL) increased ROS accumulation and induced Bcl2 phosphorylation, thereby promoting autophagic flux31. Flubendazole (a potent inducer of autophagy) activates JNK1, leading to Bcl2 phosphorylation at S70 and subsequent release of Beclin1 from Bcl2-Beclin1 complex for autophagy induction30. During mitotic arrest, sustained JNK activation dramatically increases Bcl2 phosphorylation, resulting in the inactivation of anti-apoptotic function of Bcl247. Mimicking phosphorylation of the putative mitogen-activated protein (MAP) kinase sites of Bcl2 protein (T56, T74 and S87) abolishes TNFα-induced Bcl2 degradation and apoptosis48. Bcl2 phosphorylation (T69, S70 and S87) confers resistance of CLL cells to the BH3 mimetic ABT-737 by impeding direct binding49. Together, these studies elaborated that Bcl2 phosphorylation is a pivotal mechanism for the regulation of Bcl2 functions. However, role of the phosphorylation in regulation of Bcl2 anti-apoptotic function is controversial and upstream kinase that phosphorylates Bcl2 at T69 remains elusive. In this study, we revealed that PKM2 phosphorylates Bcl2 at T69 in the mitochondria, which prevents the binding of Cul3-based E3 ligase to Bcl2 and stabilizes Bcl2, thereby inhibiting oxidative stress-induced apoptosis. Notably, Bcl2 T69 phosphorylation by PKM2 has been confirmed by in vitro kinase assay and mass spectrometry analyses.
HSP90 plays a number of important roles, such as assisting folding, intracellular transport, maintenance and degradation of proteins as well as facilitating cell signaling50. In cancer cells, it can stabilize some proteins encoded by oncogenes, such as EGFR, PI3K and AKT51,52,53. HSP90 possesses ATPase activity and induces conformational changes in client proteins, thereby causing activation of such proteins54. In this study, we demonstrate that HSP90 interacts with and isomerizes PKM2 upon oxidative stress. Isomerized PKM2 is required for Bcl2 phosphorylation and the inhibition of oxidative stress-induced apoptosis. Our finding identifies PKM2 as a new substrate of HSP90 and uncovers a novel mechanism of HSP90 regulating apoptosis via modulation of PKM2.
Our findings demonstrate that PKM2-dependent Bcl2 phosphorylation and stabilization plays an essential role in resistance of tumor cells to oxidative stress-induced apoptosis and gliomagenesis, which not only reveal a novel mechanism of PKM2-mediated ROS adaptation of tumor cells, but also provide a molecular basis for treating rapid proliferating tumors by interfering with the HSP90-PKM2-Bcl2 axis.
Materials and Methods
Materials
Rabbit monoclonal antibodies against PKM2, Bcl2 pS70, cytochrome c, AKT1, Bcl-xl, Bax, Tom20, MnSOD2 and HSP90α1, and mouse monoclonal antibodies against Bcl2 and Chk1, and polyclonal antibodies against VDAC, Cul3 and Rbx1, and mouse anti-rabbit IgG (Conformation Specific; L27A9) were obtained from Cell Signaling Technology (Danvers, MA, USA). Rabbit polyclonal antibody against Bcl2 pT69 was obtained from Thermo Fisher Scientific. Mouse monoclonal antibody against ubiquitin and GST were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit polyclonal antibody recognizing PKM2-389 (a customer antibody) was obtained from Signalway Antibody (College Park, MD, USA). Mouse monoclonal antibodies for Flag, His and tubulin, and rabbit polyclonal antibody for TIMM22 were purchased from Sigma (St. Louis, MO, USA). Hygromycin, puromycin, and G418 were purchased from EMD Biosciences (San Diego, CA, USA). PolyJet in vitro DNA transfection reagent was from Signagen Laboratories (Rockville, MD, USA). GelCode Blue Stain Reagent was obtained from Pierce (Rockford, IL, USA).
Cell culture
U87 and U251 GBM cells and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics. The protein expression and reconstitution experiments were conducted using the established stable cell lines. Transfection was performed as instructed by manufacturer.
DNA constructs and mutagenesis
PCR-amplified human PKM2 and Bcl2 were cloned into pcDNA3.1, pCDH, pCDH-Flag, pCDH-SFB, pCold I, pLOC or pGEX-KG vectors. PCR-amplified human HSP90α1 was cloned into pCMV-Flag, pCDH-Flag or pGEX-KG. Synthesized 3× PKM2 389-405 fragments or 3× Flag fragments were cloned into pCDH-Flag. pCDH-SFB-PKM2 Δ370-388, pCDH-SFB-PKM2 Δ389-405, pCDH-SFB-PKM2 406-423, pCDH-SFB-PKM2 Δ424-435, pCDH-SFB-PKM2 ILLL, pCDH-SFB-PKM2 EERR, pLOC-rPKM2, pLOC-rPKM2 ILLL, pCDH-SFB-Bcl2 T69A, pCold I-Bcl2 T69A, pCMV-Flag-HSP90α1 G97D, pCMV-Flag-HSP90α1 ΔC, pCMV-Flag-HSP90α1 MD, pCMV-Flag-HSP90α1 ΔN, pCDH-Flag-HSP90α1, pCDH-Flag-HSP90α1 1-293, pCDH-Flag-HSP90α1 1-360, pCDH-Flag-HSP90α1 1-469, pLOC-rHSP901, pCDH-Flag-rHSP90α1 G97D and pGEX-KG-HSP90α1 G97D were made using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). pLOC-rPKM2 contains nonsense mutations of C1170T, C1173T, T1174C and G1176T. pCDH-Flag-rHSP90α1 contains nonsense mutations of C2433T, A2434C and G2436T.
The pGIPZ control was generated with the control oligonucleotide 5′-GCTTCTAACACCGGAGGTCTT-3′. pGIPZ PKM2 shRNA was generated with 5′-CATCTACCACTTGCAATTA-3′ oligonucleotide targeting exon 10 of the PKM2 transcript. pGIPZ Bcl2 shRNA was generated with 5′-AGGTTCACTAAAGCAGTAG-3′ oligonucleotide targeting noncoding region of the Bcl2 transcript. pGIPZ HSP90α1 shRNA was generated with 5′-AGGATCTACAGGATGATCA-3′.
Immunofluorescence analysis
Cells were fixed and incubated with primary antibodies, Alexa Fluor dye-conjugated secondary antibodies and Hoechst 33342 according to the standard protocols. Cells were examined using a deconvolution microscope (Zeiss, Thornwood, NY, USA) with a 63-Å oil immersion objective. Axio Vision software from Zeiss was used to deconvolute Z-series images.
Subcellular fractionation analyses
U87 or U251 cells were harvested and washed three times with cold PBS. Cytosolic or mitochondrial fractions were prepared using the Mitochondria/Cytosol Fraction Kit (BioVision).
Mitochondrial subfractionation was performed as described previously55. Briefly, isolated mitochondria were resuspended in 10 μM KH2PO4 (pH 7.4) for 20 min on ice. An equal volume of iso-osmotic solution (32% sucrose, 30% glycerol, 10 mM MgCl2) was added and spun at 10 000× g for 10 min at 4 °C. The supernatant were centrifuged at 15 000× g for 1 h at 4 °C; the pellet and supernatant contained outer membrane and intermembrane space proteins, respectively. Then, the pellet from the first time centrifuge was resuspended in 10 μM KH2PO4 (pH 7.4) for 20 min on ice, and iso-osmotic solution was added, followed by centrifuge at 15 000× g for 1 h at 4 °C; the pellet and supernatant contained inner membrane and matrix proteins, respectively.
Apoptosis assay
U87 or U251 cells were treated with or without H2O2 (0.5 mM, 24 h). The cells were harvested and stained as described in the protocol of Annexin V-Cy5 Apoptosis Kit (BioVision).
ROS measurement
Total ROS in tumor tissues were measured using the Oxiselect In Vitro ROS/RNS Assay Kit (Cell Biolabs), following the manufacturer's instructions. Briefly, tumor tissues were homogenized in cold PBS solution (20 mg/ml) and centrifuged at 10 000× g for 5 min. The supernatant was collected for the assay. The fluorescence (Ex480 nm/Em530 nM) was measured using the Synergy HT Multi-Mode Microplate Reader (Bio-Tek instruments, Inc).
Caspase 3 activity assay
U87 or U251 cells were treated with or without H2O2 (0.5 mM, 24 h). The cells were harvested and washed three times with cold PBS. Caspase 3 activity was measured as described in the protocol of the Caspase-3 Activity Assay Kit (Cell Signaling Technology).
Immunoprecipitation and immunoblotting analyses
Extraction of proteins with a modified buffer from cultured cells was followed by immunoprecipitation and immunoblotting with corresponding antibodies, as described previously56.
Dot blot assay
Cell lysate was spotted on a nitrocellulose membrane (0.2 μm pore size) and then dried for several hours at room temperature. Membranes were blocked with 3% (w/v) nonfat dry milk in PBS for 30 min at room temperature and probed with the indicated antibodies for 1 h at room temperature. Membranes were washed 2 times for 3 min with PBS and subsequently incubated with a secondary antibody (HRP-conjugated goat anti-rabbit IgG,) in 3% (w/v) nonfat dry milk-PBS for 1 h at room temperature. Membranes are washed 2 times with PBS. Following the last wash, the membrane is exposed to chemiluminescence detection system (Tanon 5200 Multi) and developed using a Pierce ECL Plus Substrate (Thermo Fisher Scientific).
Purification of recombinant proteins
Wild-type and mutant GST-PKM2, His-Bcl2 and GST-HSP90α1 were expressed in bacteria and purified as described previously57. Non-tagged HSP90α1 was obtained by thrombin cleavage of GST. The purities of the recombinant proteins were verified by gel staining with Coomassie Blue.
Chaperone assay
Purified HSP90α1 (0.5 μg) and GST-PKM2 (0.5 μg) were incubated at 30 °C for 30 min with 2 mM ATP in a total volume of 20 μl, containing 40 mM HEPES-KOH (pH 7.5) and 2 mM MgCl2.
Pyruvate kinase assay
The activity of bacterially purified PKM2 WT (0.1 μg) and PKM2 ILLL (0.1 μg) toward PEP was measured with a pyruvate kinase assay (BioVision, Mountain View, CA, USA) according to the manufacturer's instruction. Data represent the mean ± SD of three independent experiments.
In vitro kinase assay
The PKM2-mediated kinase reactions were performed as described previously24. In brief, the bacterially purified recombinant PKM2 (200 ng) was incubated with Bcl2 (100 ng) with kinase buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 50 mM MgCl2, 1 mM Na3VO4, 1 mM DTT, 5% glycerol, 0.05 mM FBP, 0.5 mM PEP or 5 μCi 32P-labeled PEP) in 25 μl at 25 °C for 1 h. The reactions were terminated by the addition of SDS-PAGE loading buffer and heated to 100 °C. The reaction mixtures were then subjected to SDS-PAGE analyses. 32P-labeled PEP was prepared as described previously25. Briefly, reactions were carried out at 25 °C for 5 min. Reaction mixtures contained 50 mM HEPES, pH 7.5, 12.5 mM KF, 5 mM MgCl2, 1 mM oxaloacetate, 0.5 units PEPCK/ml and 32P-PEP. The reactions were diluted by 10-fold with water and applied to Vivapure Q columns (Sartorius) to purify 32P-labeled PEP.
Quantitative real-time PCR
Total RNA was extracted with RNeasy Plus Kit (Qiagen, Valencia, CA, USA). cDNA was prepared by a QuantiTect Reverse Transcription Kit (Qiagen) using oligonucleotide (dT) and random primers. Quantitative real-time PCR analysis was performed under the following conditions: 5 min at 95 °C followed by 40 cycles at 95 °C for 30 s, 55 °C for 40 s, and 72 °C for 1 min using an ABI Prism 7700 sequence detection system. Data were normalized to expression of a control gene (β-actin) for each experiment.
The following primer pairs were used for quantitative real-time PCR: Bcl2, 5′-GGTGGGGTCATGTGTGTGG-3′ (forward) and 5′-CGGTTCAGGTACTCAGTCATCC-3′ (reverse); β-actin, 5′-CATGTACGTTGCTATCCAGGC-3′ (forward) and 5′-CTCCTTAATGTCACGCACGAT-3′ (reverse).
Mass spectrometry analysis
An in vitro PKM2-phosphorylated recombinant Bcl2 was exhaustively acetylated with acetic anhydride and triethylamine in acetonitrile, evaporated to dryness, then resuspended in 50 mM ammonium bicarbonate buffer containing Rapigest (Waters Corp, MA, USA). The sample was heated to 95 °C for 10 min and then allowed to cool; 100 ng of sequencing-grade modified trypsin (Promega, Madison, WI, USA) was added. The digestion proceeded overnight at 37 °C and was analyzed by LC-MS/MS on an Obitrap-XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
Proteins were identified by a database search of the fragment spectra against the National SwissProt protein database (EBI) using Mascot v.2.3 (Matrix Science, London, UK) and Sequest (v.1.20) via Proteome Discoverer v.1.3 (Thermo Fisher Scientific). Phosphopeptide matches were analyzed by using PhosphoRS implemented in Proteome Discoverer and manually curated58.
Intracranial injection
We intracranially injected U87 or U251 cells (2 × 105 in 5 μl of DMEM per mouse) with depletion of endogenous PKM2 and reconstituted expression of rPKM2 WT or ILLL, or the cells with depletion of endogenous Bcl2 and reconstituted expression of rBcl2 WT or T69A into randomized 4-week-old female athymic nude mice. Intracranial injections were performed as described in a previous publication59. Seven mice per group in each experiment were included. Mice injected with U87 cells were sacrificed 40 days after implantation. U87 cells were intracranially injected into randomized athymic nude mice. After 15 days, 7 mice were sacrificed to examine tumor growth; the remaining mice were intracranially injected with the lentivirus-expressing Flag or PKM2 (389-405) peptides, and then sacrificed on day 40. The brain of each mouse was collected, fixed in 4% formaldehyde and embedded in paraffin. Tumor formation and phenotype were determined by histologic analysis of H&E-stained sections. Kaplan-Meier survival curves for the mice intracranially injected with the indicated U87 or U251 cells were performed with ten mice per group. The use of mice was in compliance with ethical regulations and was approved by the institutional review board at the Institute of Biochemistry and Cell Biology.
Bioluminescence imaging with IVIS
Mice were anaesthetized with isoflurane inhalation, and were subsequently intraperitoneally injected with 100 μl of 7.5 mg/ml D-luciferin (Xenogen). Bioluminescence imaging with a CCD camera (IVIS, Xenogen) was initiated 10 min after injection with 2 min exposure time. Bioluminescence from the region of interest was defined manually. Background was defined using a region of interest from a mouse that was not given an intraperitoneal injection of D-luciferin. All bioluminescent data were collected and analyzed using IVIS.
Immunohistochemical analysis
The tissue sections from paraffin-embedded human GBM and astrocytoma specimens were stained with antibodies. We quantitatively scored the tissue sections according to the percentage of positive cells and staining intensity, as previously defined60. We rated the intensity of staining on a scale of 0-3: 0, negative; 1, weak; 2, moderate; and 3, strong. We assigned the following proportion scores: 0 if 0% of the tumor cells showed positive staining, 1 if 0%-1% of cells were stained, 2 if 2%-10% were stained, 3 if 11%-30% were stained, 4 if 31%-70% were stained, and 5 if 71%-100% were stained. We then combined the proportion and intensity scores to obtain a total score (range, 0-8), as described previously60. Scores were compared with overall survival, defined as the time from the date of diagnosis to death or last known date of follow-up. All patients received standard adjuvant radiotherapy after surgery, followed by treatment with an alkylating agent (TMZ in most cases). The use of human brain tumor specimens and the database was approved by the Institutional Review Board at Fudan University Shanghai Cancer Center and the Institutional Review Board at First Affiliated Hospital of Wenzhou Medical College. Informed consent was obtained from all patients. Data represent the mean ± SD of 80 stained GBM specimens and 40 stained astrocytoma specimens.
Statistical analysis
We determined the significance of differences in the human glioma data using Pearson's correlation test and Student's t-test (two-tailed). P < 0.05 was considered to be significant.
Author Contributions
This study was conceived by WY; WY and JL designed the study; JL, RC, XW, PW, YZ and HG performed the experiments; FY, PW, DL and WL provided pathology assistance; CL and RZ provided technical assistance of mass spectrometry analyses; WY wrote the manuscript with comments from all authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB . Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med 2010; 49:1603–1616.
Visconti R, Grieco D . New insights on oxidative stress in cancer. Curr Opin Drug Discov Devel 2009; 12:240–245.
Waris G, Ahsan H . Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog 2006; 5:14.
Trachootham D, Alexandre J, Huang P . Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 2009; 8:579–591.
Schneider BL, Kulesz-Martin M . Destructive cycles: the role of genomic instability and adaptation in carcinogenesis. Carcinogenesis 2004; 25:2033–2044.
Martinez-Sanchez G, Giuliani A . Cellular redox status regulates hypoxia inducible factor-1 activity. Role in tumour development. J Exp Clin Cancer Res 2007; 26:39–50.
Chen EI, Hewel J, Krueger JS, et al. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res 2007; 67:1472–1486.
David CJ, Chen M, Assanah M, Canoll P, Manley JL . HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010; 463:364–368.
Noguchi T, Inoue H, Tanaka T . The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem 1986; 261:13807–13812.
Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008; 452:230–233.
Yang W, Xia Y, Cao Y, et al. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-B activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell 2012; 48:771–784.
Yang W, Zheng Y, Xia Y, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol 2012; 14:1295–1304.
Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 2011; 480:118–122.
Luo W, Hu H, Chang R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 2011; 145:732–744.
Goldberg MS, Sharp PA . Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J Exp Med 2012; 209:217–224.
Morfouace M, Lalier L, Oliver L, et al. Control of glioma cell death and differentiation by PKM2-Oct4 interaction. Cell Death Dis 2014; 5:e1036.
Meng MB, Wang HH, Guo WH, et al. Targeting pyruvate kinase M2 contributes to radiosensitivity of non-small cell lung cancer cells in vitro and in vivo. Cancer Lett 2015; 356:985–993.
Stetak A, Veress R, Ovadi J, Csermely P, Keri G, Ullrich A . Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res 2007; 67:1602–1608.
Yip KW, Reed JC . Bcl-2 family proteins and cancer. Oncogene 2008; 27:6398–6406.
Ling YH, Liebes L, Zou Y, Perez-Soler R . Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to Bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. J Biol Chem 2003; 278:33714–33723.
Hildeman DA, Mitchell T, Aronow B, Wojciechowski S, Kappler J, Marrack P . Control of Bcl-2 expression by reactive oxygen species. Proc Natl Acad Sci USA 2003; 100:15035–15040.
Liu X, Kim CN, Yang J, Jemmerson R, Wang X . Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 1996; 86:147–157.
Ku B, Liang C, Jung JU, Oh BH . Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res 2011; 21:627–641.
Yang W, Xia Y, Hawke D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 2012; 150:685–696.
Mattoo RL, Waygood EB . An enzymatic method for [32P]phosphoenolpyruvate synthesis. Anal Biochem 1983; 128:245–249.
Sato S, Fujita N, Tsuruo T . Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci USA 2000; 97:10832–10837.
Whitesell L, Lindquist SL . HSP90 and the chaperoning of cancer. Nat Rev Cancer 2005; 5:761–772.
Wen Z, Zhong Z . Darnell JE Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 1995; 82:241–250.
Peng B, Ganapathy S, Shen L, et al. Targeting Bcl-2 stability to sensitize cells harboring oncogenic ras. Oncotarget 2015; 6:22328–22337.
Alexandre J, Hu Y, Lu W, Pelicano H, Huang P . Novel action of paclitaxel against cancer cells: bystander effect mediated by reactive oxygen species. Cancer Res 2007; 67:3512–3517.
Blaser H, Dostert C, Mak TW, Brenner D . TNF and ROS crosstalk in inflammation. Trends Cell Biol 2016; 26:249–261.
Vander Heiden MG, Locasale JW, Swanson KD, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 2010; 329:1492–1499.
Yuan ZL, Guan YJ, Wang L, Wei W, Kane AB, Chin YE . Central role of the threonine residue within the p+1 loop of receptor tyrosine kinase in STAT3 constitutive phosphorylation in metastatic cancer cells. Mol Cell Biol 2004; 24:9390–9400.
Takeda K, Noguchi K, Shi W, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA 1997; 94:3801–3804.
Levine AJ, Puzio-Kuter AM . The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010; 330:1340–1344.
Bensaad K, Cheung EC, Vousden KH . Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J 2009; 28:3015–3026.
Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007; 21:2683–2710.
Liou GY, Storz P . Reactive oxygen species in cancer. Free Radic Res 2010; 44:479–496.
Anastasiou D, Poulogiannis G, Asara JM, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011; 334:1278–1283.
Lv L, Xu YP, Zhao D, et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell 2013; 52:340–352.
Gao X, Wang H, Yang JJ, Liu X, Liu ZR . Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 2012; 45:598–609.
Jiang Y, Li X, Yang W, et al. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell 2014; 53:75–87.
Jiang Y, Wang Y, Wang T, et al. PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nat Commun 2014; 5:5566.
Kim JJ, Lee SB, Park JK, Yoo YD . TNF-α-induced ROS production triggering apoptosis is directly linked to Romo1 and Bcl-X(L). Cell Death Differ 2010; 17:1420–1434.
Shirai T, Nazarewicz RR, Wallis BB, et al. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J Exp Med 2016; 213:337–354.
Li S, Swanson SK, Gogol M, et al. Serine and SAM responsive complex SESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol Cell 2015; 60:408–421.
Meshkini A, Yazdanparast R . Involvement of oxidative stress in taxol-induced apoptosis in chronic myelogenous leukemia K562 cells. Exp Toxicol Pathol 2012; 64:357–365.
Dimmeler S, Breitschopf K, Haendeler J, Zeiher AM . Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J Exp Med 1999; 189:1815–1822.
Song T, Chai G, Liu Y, Yu X, Wang Z, Zhang Z . Bcl-2 phosphorylation confers resistance on chronic lymphocytic leukaemia cells to the BH3 mimetics ABT-737, ABT-263 and ABT-199 by impeding direct binding. Br J Pharmacol 2016; 173:471–483.
Jackson SE . Hsp90: structure and function. Top Curr Chem 2013; 328:155–240.
Sawai A, Chandarlapaty S, Greulich H, et al. Inhibition of Hsp90 down-regulates mutant epidermal growth factor receptor (EGFR) expression and sensitizes EGFR mutant tumors to paclitaxel. Cancer Res 2008; 68:589–596.
Mohsin SK, Weiss HL, Gutierrez MC, et al. Neoadjuvant trastuzumab induces apoptosis in primary breast cancers. J Clin Oncol 2005; 23:2460–2468.
Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR . Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci 2006; 31:164–172.
Bukau B, Weissman J, Horwich A . Molecular chaperones and protein quality control. Cell 2006; 125:443–451.
She H, Yang Q, Shepherd K, et al. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J Clin Invest 2011; 121:930–940.
Lu Z, Liu D, Hornia A, Devonish W, Pagano M, Foster DA . Activation of protein kinase C triggers its ubiquitination and degradation. Mol Cell Biol 1998; 18:839–845.
Xia Y, Wang J, Liu TJ, Yung WK, Hunter T, Lu Z . c-Jun downregulation by HDAC3-dependent transcriptional repression promotes osmotic stress-induced cell apoptosis. Mol Cell 2007; 25:219–232.
Taus T, Kocher T, Pichler P, et al. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 2011; 10:5354–5362.
Gomez-Manzano C, Alonso MM, Yung WK, et al. Delta-24 increases the expression and activity of topoisomerase I and enhances the antiglioma effect of irinotecan. Clin Cancer Res 2006; 12:556–562.
Ji H, Wang J, Nika H, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of α-Catenin from β-Catenin and transactivation of β-Catenin. Mol Cell 2009; 36:547–559.
Acknowledgements
This work was supported by the Ministry of Science and Technology of China (2014CB943101) to WY, the National Natural Science Foundation of China (31471324 and 31422034 to WY, and 81402457 to JL), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000) to WY and the Thousand Talents Plan-Youth to WY.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Related to Figure 1. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. (PDF 509 kb)
Supplementary information, Figure S2
Related to Figure 2. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. (PDF 374 kb)
Supplementary information, Figure S3
Related to Figure 3. Immunoblotting analyses were performed with the indicated antibodies. (PDF 120 kb)
Supplementary information, Figure S4
Related to Figure 4. Immunoblotting analyses were performed with the indicated antibodies. (PDF 395 kb)
Supplementary information, Figure S5
Related to Figure 6 Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. (PDF 1629 kb)
Supplementary information, Figure S6
Related to Figure 7. Validation of the antibody specificity. (PDF 29 kb)
Supplementary information, Table S1
Related to Figure 1 293T cells were transiently transfected with SFB-PKM2 and then treated with or without H2O2 (0.5 mM, 1 h). (XLSX 12 kb)
Supplementary information, Table S2
Related to Figure 2 U87 cells that stably express SFB-PKM2 were treated with or without H2O2 (0.5 mM, 1 h). (XLSX 12 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 Unported License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Liang, J., Cao, R., Wang, X. et al. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Res 27, 329–351 (2017). https://doi.org/10.1038/cr.2016.159
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2016.159
Keywords
This article is cited by
-
LINC01852 inhibits the tumorigenesis and chemoresistance in colorectal cancer by suppressing SRSF5-mediated alternative splicing of PKM
Molecular Cancer (2024)
-
CIP2A induces PKM2 tetramer formation and oxidative phosphorylation in non-small cell lung cancer
Cell Discovery (2024)
-
The role of PKM2 in cancer progression and its structural and biological basis
Journal of Physiology and Biochemistry (2024)
-
HMGA1 regulates the mitochondrial apoptosis pathway in sepsis-induced cardiomyopathy
Cell Biochemistry and Biophysics (2024)
-
M-type pyruvate kinase 2 (PKM2) tetramerization alleviates the progression of right ventricle failure by regulating oxidative stress and mitochondrial dynamics
Journal of Translational Medicine (2023)