Abstract
Studying rare extreme forms of Alzheimer disease (AD) may prove to be a useful strategy in identifying new genes involved in monogenic determinism of AD. Amyloid precursor protein (APP), PSEN1, and PSEN2 mutations account for only 85% of autosomal dominant early-onset AD (ADEOAD) families. We hypothesised that rare copy number variants (CNVs) could be involved in ADEOAD families without mutations in known genes, as well as in rare sporadic young-onset AD cases. Using high-resolution array comparative genomic hybridisation, we assessed the presence of rare CNVs in 21 unrelated ADEOAD cases, having no alteration on known genes, and 12 sporadic AD cases, with an age of onset younger than 55 years. The analysis revealed the presence of 7 singleton CNVs (4 in ADEOAD and 3 in sporadic cases) absent in 1078 controls and 912 late-onset AD cases. Strikingly, 4 out of 7 rearrangements target genes (KLK6, SLC30A3, MEOX2, and FPR2) encoding proteins that are tightly related to amyloid-β peptide metabolism or signalling. Although these variants are individually rare and restricted to particular subgroups of patients, these findings support the causal role, in human pathology, of a set of genes coding for molecules suspected for a long time to modify Aβ metabolism or signalling, and for which animal or cellular models have already been developed.
Similar content being viewed by others
Introduction
The identification of major genes (PSEN1, PSEN2 and APP) responsible for autosomal dominant early-onset Alzheimer disease (ADEOAD, OMIM number #104300) has proved crucial to understanding the pathophysiology of the disease. It has allowed for the definition of the amyloid cascade hypothesis,1, 2 according to which accumulation of highly aggregable forms of the amyloid beta (Aβ) peptide, that results from the amyloid precursor protein (APP) proteolytic cleavage, is the primum movens of this devastating neurodegenerative disease.
ADEOAD is a rare condition with a prevalence rate estimated at 5.3 per 100 000 persons at risk.2 Mutation screening of more than 150 ADEOAD families, ascertained in France by the National Centre for early-onset AD (CNR-MAJ), had shown that mutations of known genes account for 85% of ADEOAD families.3 We hypothesised that rare copy number variants (CNVs) could be involved in ADEOAD families without mutations in known genes. Indeed, a previous analysis has shown that APP locus duplications were present in a subset of these families.4 To further explore this issue, we conducted a genome-wide screen for CNVs, using high-resolution oligonucleotide array-based comparative genomic hybridisation (a-CGH) in still unexplained ADEOAD families. In addition, as rare pathogenic CNVs often occur de novo,5 we decided to investigate by a-CGH another subgroup of patients consisting of 12 sporadic cases with particularly young disease onset (age of onset before 55 years). We reasoned that this group is likely to be enriched in de novo cases, even if, due to the pedigree structure, no parental DNA was available for confirmation. Our aim was to identify causative CNVs exclusively associated with these two disease groups and therefore, expected to be absent in a set of 1078 controls. Moreover, to assess the specificity of our findings with respect to these particular AD phenotypes, we also looked for the presence of the CNVs with exclusive association to these two disease groups in a third cohort of 912 AD cases not harbouring these extreme phenotypes. Note that in these population of unselected AD cases, previous studies have failed to detect CNV burden.6, 7
Subjects and methods
Subjects
We used a-CGH to search for pathogenic CNVs in two discovery samples corresponding to two highly selected disease groups. The first disease group included 23 unrelated probands from ADEOAD families, in which a previous screen had failed to identify any mutation on the PSEN1, PSEN2, and APP (exons 16 and 17) genes or any APP gene-dosage alteration. The pattern of inheritance was consistent with ADEOAD (ie, several AD cases with onset <60 years in at least two generations), but absence of DNA samples for a sufficient number of relatives precluded linkage analysis in these kindreds (Supplementary Figure S1). The second disease group included 12 sporadic AD cases with onset before age 55 (range 44–54 years, mean 49). These patients had been negatively screened for the same genes as ADEOAD patients. In addition, we retained only the non-carriers of an APOE4 allele to exclude a potential causal role of this potent risk factor. Patients were Caucasian of French origin, except for two patients of Italian origin and another of Moroccan origin. The 912 other AD patients (506 males, 406 females, age of onset 68.2±8.5 years, range 45–83) used to assess the specificity of our findings with respect to the two disease groups were Caucasian of French origin ascertained in Rouen (West of France). All patients fulfilled the NINCDS-ADRDA criteria for probable AD.8 The 1078 controls were mainly patients’ spouses. Ethical approval was obtained from the Paris Necker and Nord-Ouest 1 Ethics Committees, and all participants gave signed informed consent.
Oligonucleotide a-CGH
Total DNA was prepared from peripheral blood lymphocytes using the Flexigen extraction kit (Qiagen, Hilden, Germany). High-resolution a-CGH analysis was performed using the Human High-Resolution Discovery Microarray Kit 1 × 1 M (Agilent Technologies, Santa Clara, CA, USA), using standard recommended protocols, except for patient ALZ466, for whom a-CGH analysis was performed using Agilent SurePrint G3 Human 4 × 180 K catalog array (Agilent Technologies). A non-commercial genomic DNA pool of 10 control individuals was used as a reference sample. Hybridisation results were extracted with the Feature Extraction Software (10.5.1.1, Agilent Technologies, Santa Clara, CA, USA) and analyzed using Agilent's DNA-analytics software (version 4.0.81, Agilent Technologies). The data were processed using the ADM-2 algorithm, with the threshold set at 6.0 SD. A rearrangement was defined by the deviation of at least five consecutive probes.
Quantitative multiplex PCR of short fluorescent fragments
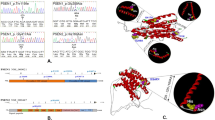
Quantitative multiplex PCR of short fluorescent fragments (QMPSF) was performed as previously described.9 Briefly, short genomic fragments (between 100 and 320 bp) of the genes subjected to CNVs were simultaneously amplified within one multiplex PCR, using 6-FAM-labelled primer pairs (Supplementary Table S1). PCR reactions were performed in 25 μl containing 100 ng of genomic DNA, 0.12–0.2 μ M of each pair of primers, and 1 unit of Thermoprime plus DNA polymerase (Abgene, Courtaboeuf, France). An amplicon of the HMBS gene was used as control. DNA fragments generated by QMPSF were separated on an ABI Prism 3100 DNA sequencer (Applied Biosystems, Courtaboeuf, France), and the resulting fluorescence profiles were analysed using the Genescan 3.7 Software (Applied Biosystems). Electropherogram of the patient (in red) was superimposed to that of a normal control (in blue) by adjusting the peaks to the same level obtained for the control amplicon (Figure 1).
a-CGH and QMPSF analyses. For each rearrangement, a-CGH analysis was confirmed using QMPSF. (a) MAGI1 partial duplication, patient ALZ 062. (b) NLRP8 duplication, patient ROU 114. (c) KLK6 duplication, patient ROU 782. (d) SLC30A3 duplication, patient ROU782. (e) FPR2 duplication, patient ALZ 466. (f) MEOX2 partial duplication, patient ROU 099. (g) SVOP partial duplication, patient EXT 068. Note that ALZ 466 a-CGH (e) analysis was performed using 4 × 180 K probe array, whereas others were performed using 1 × 1 M probe arrays, which explains the lowest probe density.
Statistical analyses
For each singleton, we computed the probability to observe no CNV in the control sample given that one CNV was observed in cases. If we assume that cases and controls were random samples from a same population in which the CNV is present at frequency q, then the number Xc of CNV alleles expected in the control sample follows a binomial distribution B(2Nc, q), with Nc being the number of controls. The probability to observe 0 CNV allele is then P0=P (Xc=0)=(1−q)2Nc. An estimate of q with the corresponding 95% confidence interval (CI) can be obtained from the sample of Nd cases: E(q)=1/2Nd, with Nd being the number of cases and V(q)=(1/2Nd)*(1−(1/2Nd))/2Nd.
Results
On average, 45 CNVs (27 deletions and 18 duplications) were detected per individual. Their size ranged from 2.5 to 7.9 Mb, with a median size of 29.8 kb. CNVs present in the database of genomic variants (http://projects.tcag.ca/variation/), excluding BAC array-based studies, or CNVs present in nongenic or intronic regions were filtered out. At this stage, 18 CNVs (5 deletions and 13 duplications) were retained. These CNVs were then all confirmed by targeted QMPSF analyses, and their frequencies were determined in a sample of 1078 controls using QMPSF. CNVs present in the control sample were then excluded. At the completion of this analysis, four singleton CNVs, absent in controls, were retained in the ADEOAD group and three in the sporadic cases group (Table 1). For each CNV observed once among the 21 ADEOAD cases (q=0.024; 95% CI=(0.001; 0.123)), the probability to have not observed it in the 1078 controls is 2.73 × 10−23 and does not exceed 0.07 at the lower bound of the CI. For each CNV observed once among the 12 sporadic cases (q=0.042, 95% CI=90.002; 0.202)), the probability is 1.41 × 10−40 and 0.01 at the lower bound of the CI.
All retained CNVs were duplications, with a size ranging from 15 to 362 kb (median size of 103 kb). In index cases of ADEOAD families, we observed (i) a duplication including NLRP8, as well as a large part of the NLRP5 gene, (ii) a partial duplication of MAGI1, (iii) a duplication of KLK6, and (iv) a duplication located upstream of the MEOX2 gene and including exon 1 of the gene. In sporadic early-onset AD cases, we observed (v) a partial duplication of SVOP and USP30 in a patient with onset at age 45, (vi) a large duplication including part of the HAS1 gene, as well as the FPR1, FPR2, and FPR3 genes, in a patient with onset at age 47 and (vii) a duplication including SLC30A3, DNAJC5G, and part of the TRIM54 gene, in a patient with onset at age 48 (Figure 1). QMPSF analysis showed that these 7 CNVs were not present in the 912 other AD cases. The only positive QMPSF signal detected in these patients corresponded to the gain of one copy of an NLRP8 amplicon in a subject with onset at age 80 years. Subsequent a-CGH analysis revealed that this patient bore a small duplication of NLRP8 exons 1–3. This rearrangement, which was reported in the database of genomic variants, was thus only partially overlapping with that found in the ADEOAD case (Supplementary Figure S2). More generally, note that other partially overlapping rearrangements, different from those found in our patients and including short intronic indels, have been reported at some of these loci in the Database of Genomic variants (Supplementary Table S2).
Discussion
Three CNVs contained genes whose relationship with AD pathology, if any, remains elusive. MAGI1 (3p14.1) and SVOP (12q24.11) encode synaptic proteins currently without any known link to AD pathological processes. USP30 (12q24.11) encodes a deubiquitinating enzyme present in the mitochondrial outer membrane. NLRP8 (19q13.42) encodes a member of the cytosolic Nod-like receptors that have a role in regulation of innate and adaptative immunity. Although the exact functions and signalling pathways of NLRP8 remain obscure, it should be noted that several NLR proteins are components of inflammasome, a multiprotein complex, which is activated in AD brains by Aβ oligomers.10 Thus, a possible link between NLRP8, inflammation, and AD can be envisioned.
The four remaining CNVs also contain several genes with currently no known relevance to AD pathophysiology: HAS1 (19q13.33) encodes a hyaluronan synthase involved in the biosynthesis of glycosaminoglycans of extracellular matrix; DNAJC5G (2p23.3) is an uncharacterized homologue of the hsp40 chaperon; and TRIM54 (2p23.3) encodes a muscle-specific ring finger protein not expressed in the brain. Besides these genes, however, four other genes included in these CNVs encode proteins whose function is firmly related to the Aβ biological network:
-
KLK6 (19q13.33) encodes neurosin, a secreted kallikrein-like protease localised in senile plaques and neurofibrillary tangles of AD brains.11 Cells cotransfected with APP and KLK6 release an abundance of amyloidergic N-truncated Aβ peptides and concomitantly, show a reduction of production of non-amyloidergic species resulting from alpha secretase activity.12
-
MEOX2 (7p21.1) encodes a homeobox protein, known to be a regulator of vascular differentiation, whose expression is low in AD. Restoring the expression of MEOX2 in brain endothelial cells of AD patients stimulates angiogenesis and increases the level of LRP, a major Aβ clearance receptor, at the blood-brain barrier. Conversely, in mice, deletion of MEOX2 results in brain capillary deficit and impaired Aβ efflux caused by reduced LRP levels.13
-
SLC30A3 (2p23.3) encodes the ZnT3 synaptic vesicle zinc transporter responsible for neuronal Zn2+ export into the synaptic space or in perivascular spaces and leptomeningal vessels. Zn2+ binds directly to Aβ and promotes its aggregation in senile plaques or in cerebral amyloid angiopathy (CAA). In a transgenic mouse model of AD (Tg2576), a dramatic reduction of CAA occurs after targeted disruption of SLC30A3, supporting the view that synaptic ZnT3 activity promotes CAA by raising exchangeable Zn2+ levels.14 Recently, it has also been shown that ZnT3 activity is critical for Aβ oligomers synaptic targeting.15
-
The FPR genes (19q13.33) encode formyl peptide receptors, which are G protein-coupled receptors. FPR1 is thought to mediate the response of phagocytic cells to invasion of the host by microorganisms and FPR3 is characterized by its specific expression in monocytes and dendritic cells. FPR2, which is expressed in brain phagocytes, is a functional receptor used by Aβ-42 to chemoattract and activate mononuclear phagocytic cells.16, 17 Neuroblastoma cell lines expressing FPR2 exhibit increased Aβ-42-induced cellular death. Interestingly, the neuroprotective peptide Humanin that protects cells from damage by Aβ-42 also binds to FPR2 without inducing apoptotic death. It has been suggested that Humanin exerts its neuroprotective effect by competitively inhibiting the access of FPR2 to Aβ-42.18
In complex diseases, focusing on subgroups of patients with an extreme phenotype is a useful strategy in revealing rare genetic causal factors.19 Using this approach, we have identified seven CNVs, exclusive to ADEOAD or sporadic cases with APOE 33 genotype and disease onset <55 years. None of these CNVs were retrieved in other AD patients, demonstrating a strong specificity for these extreme phenotypes. For each CNV, sequence analysis of the boundary regions did not reveal any interstitial duplication or Alu sequences, suggesting that these rearrangements are not mediated by a non-homologous allelic rearrangement mechanism, which is consistent with their lack of recurrence.
We cannot exclude that the CNVs found exclusively in our two selected disease groups correspond to private non-pathogenic variations. However, arguing strongly against this interpretation is the fact that among the seven CNVs retained at the final stage from this large genome scan, four included genes involved in Aβ-related biological pathways and a fifth was possibly involved in the same network. If we consider these CNVs as ultra low-frequency benign variants, it seems highly unlikely that at least four of the seven haphazardly impact genes strongly related to Aβ metabolism or signalling. Three of these genes (KLK6, SLC30A3, and FPR2) are entirely duplicated. The direction of the observed gene dosage alteration is consistent with overexpression of the protein and is in agreement with the deleterious effect predicted from animal or cellular models. The MEOX2 gene is affected by a partial duplication at the 5′-end of the gene. Although the functional consequence of this kind of rearrangement is more difficult to predict, it can produce non-functional fusion transcripts or alter gene expression by disrupting regulatory elements located in this region, in accordance with the animal model, predicting a deleterious loss of function.13
In conclusion, focusing on rare DNA alterations found exclusively in highly selected populations of AD patients, we have shown that a set of genes coding for molecules long suspected of modifying Aβ metabolism (and for which animal or cellular models had already been developed), most likely has a role in AD pathophysiology. These results provide novel support for the amyloid cascade hypothesis.1 However, it should be stressed that definitive proof of the causal involvement of these genes requires segregation analysis, which was not possible in our families, or identification of similar alterations in other cases with these extreme phenotypes. Nevertheless, particular attention should be devoted to these genes in forthcoming next-generation sequencing studies.
References
Hardy J, Selkoe DJ : The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002; 297: 353–356.
Campion D, Dumanchin C, Hannequin D et al: Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet 1999; 65: 664–670.
Raux G, Guyant-Marechal L, Martin C et al: Molecular diagnosis of autosomal dominant early onset Alzheimer's disease: an update. J Med Genet 2005; 42: 793–795.
Rovelet-Lecrux A, Hannequin D, Raux G et al: APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 2006; 38: 24–26.
Lupski JR : Genomic rearrangements and sporadic disease. Nat Genet 2007; 39: S43–S47.
Heinzen EL, Need AC, Hayden KM et al: Genome-wide scan of copy number variation in late-onset Alzheimer's disease. J Alzheimers Dis 2010; 19: 69–77.
Swaminathan S, Kim S, Shen L et al: Genomic copy number analysis in Alzheimer's disease and mild cognitive impairment: an ADNI study. Int J Alzheimers Dis 2011; 2011: 729478.
McKhann G, Drachman D, Folstein M et al: Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 1984; 34: 939–944.
Charbonnier F, Raux G, Wang Q et al: Detection of exon deletions and duplications of the mismatch repair genes in hereditary nonpolyposis colorectal cancer families using multiplex polymerase chain reaction of short fluorescent fragments. Cancer Res 2000; 60: 2760–2763.
Salminen A, Ojala J, Suuronen T, Kaarniranta K, Kauppinen A : Amyloid-beta oligomers set fire to inflammasomes and induce Alzheimer's pathology. J Cell Mol Med 2008; 12: 2255–2262.
Ogawa K, Yamada T, Tsujioka Y et al: Localization of a novel type trypsin-like serine protease, neurosin, in brain tissues of Alzheimer's disease and Parkinson's disease. Psychiatry Clin Neurosci 2000; 54: 419–426.
Little SP, Dixon EP, Norris F et al: Zyme, a novel and potentially amyloidogenic enzyme cDNA isolated from Alzheimer's disease brain. J Biol Chem 1997; 272: 25135–25142.
Wu Z, Guo H, Chow N et al: Role of the MEOX2 homeobox gene in neurovascular dysfunction in Alzheimer disease. Nat Med 2005; 11: 959–965.
Friedlich AL, Lee JY, van Groen T et al: Neuronal zinc exchange with the blood vessel wall promotes cerebral amyloid angiopathy in an animal model of Alzheimer's disease. J Neurosci 2004; 24: 3453–3459.
Deshpande A, Kawai H, Metherate R, Glabe CG, Busciglio J : A role for synaptic zinc in activity-dependent Abeta oligomer formation and accumulation at excitatory synapses. J Neurosci 2009; 29: 4004–4015.
Iribarren P, Zhou Y, Hu J, Le Y, Wang JM : Role of formyl peptide receptor-like 1 (FPRL1/FPR2) in mononuclear phagocyte responses in Alzheimer disease. Immunol Res 2005; 31: 165–176.
Tiffany HL, Lavigne MC, Cui YH et al: Amyloid-beta induces chemotaxis and oxidant stress by acting at formylpeptide receptor 2, a G protein-coupled receptor expressed in phagocytes and brain. J Biol Chem 2001; 276: 23645–23652.
Ying G, Iribarren P, Zhou Y et al: Humanin, a newly identified neuroprotective factor, uses the G protein-coupled formylpeptide receptor-like-1 as a functional receptor. J Immunol 2004; 172: 7078–7085.
Cirulli ET, Goldstein DB : Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010; 11: 415–425.
Acknowledgements
We are indebted to the banque d’ADN et de cellules Pitié Salpêtrière. We thank Dr Mario Tosi and Tracey Avequin for critical reading of the manuscript and Emmanuelle Genin for statistical support. This study was funded by PHRC GMAJ 2008/067 (Rouen University Hospital).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Appendix
Appendix
The investigators of the French GMAJ project include Didier Hannequin, Dominique Campion, Olivier Martinaud, Lucie Guyant-Maréchal and David Wallon (Centre Hospitalo Universitaire (CHU), Rouen); Olivier Godefroy and Candice Picard (CHU Amiens); Frédérique Etcharry-Bouyx (CHU Angers); Eric Berger (CHU Besancon); Jean-Francois Dartigues and Sophie Auriacombe (CHU Bordeaux); Vincent de la Sayette (CHU Caen); Francois Sellal (CH Colmar); Olivier Rouaud and Christelle Thauvin (CHU Dijon); Olivier Moreaud (CHU Grenoble); Stéphanie Bombois, Adeline Rollin-Sillaire, Marie-Anne Mackowiak and Florence Pasquier (CHU Lille); Isabelle Roullet-Solignac and Alain Vighetto (CHU Lyon); Mira Didic, Olivier Félician and Mathieu Ceccaldi (CHU Marseille); Audrey Gabelle and Jacques Touchon (CHU Montpellier); Martine Vercelletto and Claire Boutoleau-Bretonnière (CHU Nantes); Pierre Labauge and Giovanni Castelnovo (CHU Nimes); Claire Paquet and Jacques Hugon (CHU Lariboisière); Agnès Michon, Isabelle Le Ber and Bruno Dubois (CHU La Salpêtrière, Paris); Catherine Thomas-Antérion (CHU Saint-Etienne); Frédéric Blanc and Christine Tranchant (CHU Strasbourg); Jérémie Pariente, Michèle Puel and Jean-Francois Demonet (CHU Toulouse); Caroline Hommet and Karl Mondon (CHU Tours); Hélène Mollion and Bernard Croisile (CMRR CHU Lyon); Mathilde Sauvée (CHU Nancy); Gaelle Godenèche and Foucauld De Boisgueheneuc (CHU Poitiers).
Rights and permissions
About this article
Cite this article
Rovelet-Lecrux, A., Legallic, S., Wallon, D. et al. A genome-wide study reveals rare CNVs exclusive to extreme phenotypes of Alzheimer disease. Eur J Hum Genet 20, 613–617 (2012). https://doi.org/10.1038/ejhg.2011.225
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.225
Keywords
This article is cited by
-
Genetic Phenotypes of Alzheimer’s Disease: Mechanisms and Potential Therapy
Phenomics (2023)
-
Dissecting the clinical heterogeneity of early-onset Alzheimer’s disease
Molecular Psychiatry (2022)
-
Dissection of the polygenic architecture of neuronal Aβ production using a large sample of individual iPSC lines derived from Alzheimer’s disease patients
Nature Aging (2022)
-
The role of de novo mutations in adult-onset neurodegenerative disorders
Acta Neuropathologica (2019)
-
NLRs as Helpline in the Brain: Mechanisms and Therapeutic Implications
Molecular Neurobiology (2018)