Abstract
Transcobalamin II (TC II) is a plasma transport protein for cobalamin. TC II deficiency can lead to infant megaloblastic anemia, failure to thrive and to neurological complications. This report describes the genetic work-up of three patients who presented in early infancy. Initially, genomic investigations did not reveal the definite genetic diagnosis in the two index patients. However, analysis of cDNA from skin fibroblasts revealed a homozygous deletion of exon 7 of the TC II gene caused by the mutation c.940+303_c.1106+746del2152insCTGG (r.941_1105del; p.fs326X) in one patient. The other patients were siblings and both affected by an insertion of 87 bp on the transcript which was caused by the homozygous mutation c.580+624A>T (r.580ins87; p.fs209X). Additional experiments showed that cDNA from lymphocytes could have been used also for the genetic work-up. This report shows that the use of cDNA from skin fibroblasts or peripheral lymphocytes facilitates genetic investigations of suspected TC II deficiency and helps to avoid false-negative DNA analysis.
Similar content being viewed by others
Introduction
Transcobalamin II (TCN2; TC II) is a plasma protein that is essential for the absorption, transport and cellular uptake of cobalamin (Cbl, vitamin B12).1, 2 Cbl is synthesized only by lower organisms and not by plants and animals. It accumulates in animals so that animal products are the only source of Cbl in the human diet. Intrinsic factor-bound Cbl is recognized by ileal receptor proteins, cubilin and megalin, and enters the enterocytes through endocytosis. In ileal enterocytes, Cbl is dissolved from the intrinsic factor and is bound to TC II before release into the blood system and distribution to target tissues.3, 4
Transcobalamin II deficiency (TC II deficiency, MIM no. 275350) leads to a lack of intracellular Cbl, which manifests mainly in early infancy. Symptoms include megaloblastic anemia and pancytopenia, vomiting, diarrhea and early-onset of failure to thrive, repeated infections, and mental retardation.4, 5, 6, 7, 8 The disorder is inherited in an autosomal-recessive trait and was first described in 1971 in two siblings with megaloblastic anemia and normal levels of serum Cbl.9
Diagnosis can either be achieved by the measurement of serum transcobalamin by an enzyme-linked immunosorbent assay method10 or by genetic testing. Serum Cbl levels cannot serve as a diagnostic tool because they show as normal and most of serum Cbl is bound to haptocorrin (or R-binder, formerly named transcobalamin I), a glycoprotein whose function is not fully understood.4, 5
In this study, the genetic work-up of two patients with typical early-onset TC II deficiency is reported. Exon amplification and sequencing of the TCII gene was uninformative with regard to the definite genetic background in both patients. Skin fibroblast-derived transcript analysis elucidated the genotypes. Additional experiments showed that lymphocytes could have been used as a source of the cDNA. Thus, transcript investigations using peripheral lymphocytes or skin fibroblasts can be regarded as an economical and straightforward approach for mutation screening in TC II deficiency.
Materials and methods
Patients
Patient 1 is the daughter of consanguineous Lebanese parents born at term (birth weight 3190 g, length 50 cm) after an uneventful pregnancy. Symptoms started at the age of 3 weeks with diarrhea, vomiting, failure to thrive, as well as floppiness and muscular hypotonia. Investigations revealed pancytopenia (white blood cells 5100/μl, hemoglobin 5.7 g per 100 ml, MCV 85 fl, platelets 72.000/μl, reticulocytes 6%), normal levels of serum Cbl (477 pg ml−1), folic acid (9.3 ng ml−1) and distinct changes of plasma amino acids: homocysteine 25.4 μmol l−1 (reference <8), total cysteine 87 μmol l−1 (reference 200–460), methionine 11 μmol l−1 (reference 20–40), cystine 4 μmol l−1 (reference 20–60). Urine organic acids yielded a moderately elevated excretion of methylmalonic acid. Bone marrow aspiration showed a hypercellular bone marrow and as main pathology, a markedly altered erythropoiesis showing megaloblastic transformations and anomalies of nuclei forms suggestive of a Cbl or folic acid deficiency. TC II levels in serum were not established. Oral substitution of 15 mg folinic acid per day and intramuscular application of 1.5 mg hydroxycobalamin three times a week led to clinical and hematological recovery within 2 months. Follow-up investigations at age 18 months showed a normal mental development but significant motor deficits (Münchener Funktionelle Entwicklungsdiagnostik).
Patients 2 and 3 are offspring of consanguineous Turkish parents. The elder sister (patient 2 in this report) became symptomatic at the age of 6 weeks with failure to thrive and pancytopenia (white blood cells 580/μl, hemoglobin 6.3 g per 100 ml, platelets 119.000/μl). Bone marrow aspiration showed a reduced erythropoiesis and thrombopoiesis but an increased granulopoiesis. At 8 weeks, urine organic acid analysis revealed massively elevated excretion of methylmalonic acid, methylcitrate and 3-OH-proprionate. As with patient 1, TC II levels in serum were not established before initiation of treatment. Application of oral folic acid and intramuscular hydroxycobalamin (at start 1 mg daily) led to the normalization of clinical symptoms and urine metabolites within 3 weeks as well as hematological abnormalities within 3 months. Symptoms in patient 3 started at week 6 of life with vomiting and failure to thrive. The laboratory investigations showed an anemia (hemoglobin 7.5 g per 100 ml), hyperhomocysteinemia (149 μmol l−1) with a low normal methionine (15 μmol l−1) and a massively elevated excretion of methylmalonic acid in urine. Serum Cbl was normal (864 pg ml−1). On the basis of family history, an oral supplementation of folic acid and parenteral hydroxycobalamin (at start 1 mg daily) was initiated, which led to a full recovery within 8 weeks. A follow-up at age of 9 and 11 years, respectively, showed a normal psychomotor development in patients 2 and 3.
For molecular genetic diagnostics, blood samples of patients were taken and skin fibroblasts from patients 1 and 2 were put into culture after informed consent of parents was obtained. For the purpose of lymphocyte isolation and subsequent cDNA synthesis, blood samples of two healthy German individuals were collected after written consent for the genetic study was obtained. In addition, DNA samples of 50 healthy German individuals were used as controls after written consent for the use of the samples for scientific purposes was obtained.
DNA isolation and cDNA synthesis
Genomic DNA was extracted from patient's whole blood cells by a standard method (QIAamp blood kit, Qiagen, Hilden, Germany).
Total RNA was isolated from cultured fibroblasts of the patients (RNeasy Mini Kit, Invitrogen, Groningen, The Netherlands) to synthesize single-stranded cDNA according to the manufacturer's recommendations (First Strand cDNA Synthesis Kit for reverse transcription-PCR, Boehringer Mannheim, Mannheim, Germany). cDNA from peripheral lymphocytes of two healthy controls was produced by the same method.
Mutation analysis using DNA and cDNA
Primers were designed binding in the flanking intronic regions of each of the nine coding exons (see Table 1). PCR amplification was performed at an annealing temperature of 64 °C using Taq-Polymerase (Boehringer Mannheim) before direct sequencing of PCR products using the ABI Prism 3700 DNA Sequencer (Applied Biosystems, Foster City, CA, USA).
A full-length PCR of the TC II transcript was performed at an annealing temperature of 62 °C (expected size 1458 bp; primers given in Table 1). This was later repeated using cDNA derived from uncultured peripheral lymphocytes obtained from control individuals. PCR products were purified following agarose gel electrophoresis and subjected to direct sequencing using the ABI Prism 3700 DNA Sequencer (Applied Biosystems).
For characterization of the exact size of the assumed deletion in patient 1, intronic primers for the region of interest were used for PCR amplification of DNA at an annealing temperature of 65 °C (primer-forward 5′-CAATCTGCTGCGCACCCATT-3′, primer-reverse 5′-CAGGTGTTAGCCATCACACTC-3′). PCR products were directly sequenced as above. For the amplification of the region surrounding the inserted sequence of intron 4 in patient 2, an annealing temperature of 64 °C was used for the forward-primer for exon 4 and reverse-primer for exon 5, respectively.
Results
Direct sequencing of the nine coding exons of the TC II gene, including the flanking intronic regions, did not reveal the definite genotype in either of the two index patients.
In patient 1, PCR amplification of exon 7 did not yield a product indicating a possible deletion in this genomic region. On the basis of these results and on the strong clinical and biochemical suspicion, it was decided to proceed with RNA diagnostics.
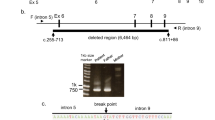
Reverse transcription-PCR of skin fibroblast-derived cDNA yielded full-length TC II fragments that were about 200 bp shorter in size in patient 1 and about 80 bp longer in patient 2 when compared with controls. Direct sequencing of full-length TC II reverse transcription-PCR products revealed a homozygous deletion of exon 7 of the TC II transcript in cDNA derived from patient 1. For breakpoint characterization, PCR amplification with a forward primer located in intron 6 and a reverse primer in intron 7 was performed and yielded products that were about 2000 bp shorter than healthy controls. Direct sequencing indicated breakpoints at position c.940+303 in intron 6 and at position c.1106+746 in intron 7, respectively. This resulted in a genomic deletion of 2152 bp and an insertion of CTGG (c.940+303_c.1106+746del2152insCTGG) as well as a subsequent loss of exon 7 (r.941_1105del) in a homozygous state (Figure 1a). The loss of exon 7 affects the transcript with a frameshift and results in a premature stop of translation at codon 326 (fs326X). Both parents were shown to be heterozygous carriers of the respective mutation.
Illustration of the effect of the mutations both on genomic DNA and on cDNA. Gray boxes indicate coding exons with numbering given. Light gray triangles display the result of the genomic mutation on cDNA. (a) Dashed white box indicates exon 7, which was found to be deleted in patient 1. Vertical arrows indicate location of breakpoints of the deletion. (b) Vertical arrow indicates location of the intronic nucleotide exchange leading to creation of a novel intronic donor splice site. ag, cryptic intronic acceptor splice site; gt, novel intronic donor splice site; bp, base pairs.
In patient 2, sequencing of the full-length TC II transcript revealed a homozygous insertion of 87 bp between exons 4 and 5 with the inserted sequence being identical to a sequence in intron 4. PCR amplification and sequencing of part of intron 4 revealed the mutation c.580+624A>T in a homozygous state. This mutation generates a new donor splice site leading to activation of a cryptic acceptor splice site and results in the insertion of the transcript (Figure 1b) and a premature stop of translation within the inserted region (r.580ins87bp; fs209X). Direct sequencing of 100 control chromosomes did not reveal the nucleotide exchange c.580+624A>T. This mutation was also confirmed in DNA from patient 3 in a homozygous state while both parents were found to be heterozygous for the respective mutation.
In addition, cDNA derived from uncultured peripheral lymphocytes of controls was used for amplification of the full-length TC II transcript. This yielded a specific reverse transcription-PCR product that was successfully subjected to direct sequencing indicating that transcript analysis using peripheral blood cells is feasible.
Discussion
In this paper, we have added three patients to the short list of reported patients suffering from TC II deficiency. The results of the patients reported in this paper follow published characteristics4 with respect to the clinical course and most laboratory results. However, the finding of the hypercellular bone marrow in patient 1 is unusual and might indicate a possible variability of this sign in TC II deficiency. As well documented in all reports, supplementation of Cbl is an effective treatment for this condition. This is noteworthy, given the lack of its transporter in blood. The feasibility of this treatment is not fully understood. However, it might be explained by some level of residual activity of the mutated protein or by an alternative transport of Cbl in blood by binding to haptocorrin, albumin or to other proteins. It should be noted that the route of administration of Cbl is still under discussion. Although the patients reported here were treated by parenteral Cbl, oral treatment, if carefully monitored,4, 11 might be similarly effective.
Although known as a clinical entity since 1971 and despite description of first mutations in the TC II gene as early as 19949, 12, there are only a total of six mutations found in eight patients reported in the literature.7, 12, 13, 14 There is considerable genetic heterogeneity of TC II deficiency comprising nonsense mutations,12 deletions,13 RNA editing7 and a mutation affecting a splice site14 (details of mutations are provided in Table 2).
The human TC II gene on chromosome 22q11.2-qter has been fully cloned and characterized in 1995 allowing for genetic investigations based on DNA amplification.15, 16 However, the patients described in this study were not fully informative when the nine coding exons, including flanking intronic regions, of the TC II gene were sequenced. On the basis of clinical and biochemical strong impression of TC II deficiency as being the underlying disorder in the patients, we proceeded by skin fibroblasts-derived RNA investigations to confirm the suspected diagnosis. It has yet to be emphasized that serum studies of transcobalamin could have been performed to confirm the suspected diagnosis.
According to DNA and RNA studies, patient 1 was found to be affected by a homozygous deletion of 2152 bp and an insertion of four nucleotides (CTGG) resulting in the loss of exon 7 of the TC II gene. This mutation causes a frameshift with a premature stop of translation at codon 326 and can, therefore, be considered as disease causing. Patients 2 and 3 are affected by an intronic nucleotide exchange. This creates a novel intronic donor splice site leading to the use of a cryptic intronic acceptor splice site and a consecutive insertion of 87 bp into the TC II transcript. As the inserted fragment carries an in-frame stop codon at position 209, this mutation results also in a truncated TC II protein and can be regarded as disease causing. This intronic mutation adds to the hitherto described genetic heterogeneity of TC II deficiency as mentioned above.
To facilitate future genetic investigations, in addition we investigated RNA from control lymphocytes as an alternative, which allows waiving skin biopsies. Hereby, the feasibility of this source of material for genetic work-up of TC II deficiency could be demonstrated.
In conclusion, this study reports on three patients with TC II deficiency in which RNA-based molecular genetics but not exon amplification and sequencing of DNA led to the definite genetic diagnosis. We show that lymphocytes can be used for full-length cDNA analysis to complement genomic investigations facilitating genetic diagnostics of TC II deficiency and to avoid false-negative DNA investigations.
Accession codes
References
Hall, C. A. & Finkler, A. E. Function of transcobalamin II: a B-12 binding protein in human plasma. Proc. Soc. Exp. Biol. Med. 123, 55–58 (1966).
Jacob, E., Baker, S. J. & Herbert, V. Vitamin B12-binding proteins. Physiol. Rev. 60, 918–960 (1980).
Rothenberg, S. P., Weiss, J. P. & Cotter, R. Formation of transcobalamin II-vitamin B12 complex by guinea-pig ileal mucosa in organ culture after in vivo incubation with intrinsic factor-vitamin B12. Br. J. Haematol. 40, 401–414 (1978).
Rosenblatt, D. S. & Fenton, W. A. in The Metabolic and Molecular Bases of Inherited Disease (eds, Scriver, C.R., Beaudet, A.L., Valle, D. and Sly, W.S.) 3915–3916 (McGraw Hill, New York, 2001).
Frater-Schröder, M. Genetic patterns of transcobalamin II and the relationships with congenital defects. Mol. Cell Biochem. 56, 5–31 (1983).
Hall, C. A. The neurologic aspects of transcobalamin II deficiency. Br. J. Haematol. 80, 117–120 (1992).
Qian, L., Quadros, E. V., Regec, A., Zittoun, J. & Rothenberg, S. P. Congenital transcobalamin II deficiency due to errors in RNA editing. Blood Cells Mol. Dis. 28, 134–142 (2002).
Scott, C. R., Hakami, N., Teng, C. C. & Sagerson, R. N. Hereditary transcobalamin II deficiency: the role of transcobalamin II in vitamin B 12-mediated reactions. J. Pediatr. 81, 1106–1111 (1972).
Hakami, N., Neiman, P. E., Canellos, G. P. & Lazerson, J. Neonatal megaloblastic anemia due to inherited transcobalamin II deficiency in two siblings. N. Engl. J. Med. 285, 1163–1170 (1971).
Nexo, E., Christensen, A. L., Petersen, T. E. & Fedosov, S. N. Measurement of transcobalamin by ELISA. Clin. Chem. 46, 1643–1649 (2000).
Bibi, H., Gelman-Kohan, Z., Baumgartner, E. R. & Rosenblatt, D. S. Transcobalamin II defciency with methylmalonic aciduria in three sisters. J. Inher. Metab. Dis. 22, 765–772 (1999).
Li, N., Rosenblatt, D. S. & Seetharam, B. Nonsense mutations in human transcobalamin II deficiency. Biochem. Biophys. Res. Commun. 204, 1111–1118 (1994a).
Li, N., Rosenblatt, D. S., Kamen, B. A., Seetharam, S. & Seetharam, B. Identification of two mutant alleles of transcobalamin II in an affected family. Hum. Mol. Genet. 3, 1835–1840 (1994b).
Namour, F., Helfer, A. C., Quadros, E. V., Alberto, J. M., Bibi, H. M., Orning, L. et al. Transcobalamin deficiency due to activation of an intra exonic cryptic splice site. Br. J. Haematol. 123, 915–920 (2003).
Li, N., Seetharam, S. & Seetharam, B. Genomic structure of human transcobalamin II: comparison to human intrinsic factor and transcobalamin I. Biochem. Biophys. Res. Commun. 208, 756–764 (1995).
Regec, A., Quadros, E. V., Platica, O. & Rothenberg, S. P. The cloning and characterization of the human transcobalamin II gene. Blood 85, 2711–2719 (1995).
Acknowledgements
We thank Dr R Brackmann (Herford) and Dr P Navratil (Coesfeld) for referring patients for further investigations. In addition, the technical assistance of Ingrid DuChesne and Ilka Neumann is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Häberle, J., Pauli, S., Berning, C. et al. TC II deficiency: avoidance of false-negative molecular genetics by RNA-based investigations. J Hum Genet 54, 331–334 (2009). https://doi.org/10.1038/jhg.2009.34
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.34
Keywords
This article is cited by
-
Identification of transcobalamin deficiency with two novel mutations in the TCN2 gene in a Chinese girl with abnormal immunity: a case report
BMC Pediatrics (2020)
-
A review of the diverse genetic disorders in the Lebanese population: highlighting the urgency for community genetic services
Journal of Community Genetics (2015)
-
A novel mutation of the transcobalamin II gene in an infant presenting with hemophagocytic lymphohistiocytosis
International Journal of Hematology (2014)
-
Update on transcobalamin deficiency: clinical presentation, treatment and outcome
Journal of Inherited Metabolic Disease (2014)
-
Transcobalamin deficiency caused by compound heterozygosity for two novel mutations in the TCN2 gene: a study of two affected siblings, their brother, and their parents
Journal of Inherited Metabolic Disease (2010)