Abstract
The male genetic landscape of the territory currently known as Tunisia is hampered by the scarcity of data, especially from cosmopolitan areas such as the coastal city of Sousse. In order to alleviate this lacuna, 220 males from Sousse were examined, for the first time, for more than 50 Y-chromosome single-nucleotide polymorphisms (Y-SNPs) markers and compared with 3099 individuals from key geographically targeted locations in North Africa, Europe and the Near East. The paternal lineages observed belong to a common set of Y haplogroups previously described in North Africa. In addition to the prominent autochthonous North African E-M81 haplogroup which is exclusively represented by its subclade E-M183 (44.55% of Y-chromosomes), a number of Near Eastern Neolithic lineages including E-M78, J-M267 and J-M172 account for 39% of the Y-chromosomes detected. Principal component analysis based on haplogroup frequencies, multidimensional scaling based on Rst genetic distances and analyses of molecular variance using both Y-chromosome short tandem repeat haplotypes and Y-SNP haplogroup data revealed that the Tunisian and North African groups, as a whole, are intra- and inter-specific diverse with Sousse being highly heterogeneous.
Similar content being viewed by others
Introduction
Located at the northernmost bulge of Africa between Europe and the Middle East, Tunisia is a Mediterranean country bounded on the north and the east by an extensive coastline (1300 km).1, 2 From this strategic location, Tunisia has been a crossroads of human migrations since the Paleolithic period, leading to population admixture that has increased during more than 3000 years of history.3 As a result, modern Tunisian populations display high intra- and inter-specific genetic diversity. Although explored by different disciplines, the peopling of North Africa, and Tunisia particularly, remain a subject of continuous debate. Approximately 8000 years before the common era, Tunisia was characterized by the Capsian culture, which developed in situ in the Maghreb region of Northwest Africa and subsequently experienced a Neolithic transition in its later stages.4, 5
In historical times, because of its location on the main maritime roads of the Mediterranean, Tunisia has been settled successively by many diversed populations including Phoenicians, Romans, Vandals and Byzantines. By the end of the seventh century, Muslim armies from the Arabian Peninsula invaded North Africa and reached the region known today as Tunisia. In the late tenth century, Tunisia also experience an important movement of Arab populations, mostly Bedouin.6, 7 Therefore, it is likely that migrations and admixture processes might have played a pivotal role in shaping the peopling of Tunisia, particularly in coastal cities. In addition, because of its numerous small, isolated indigenous populations, Tunisia represents an interesting region to explore inter population relationships. Indeed, European, Near Eastern and sub-Saharan contributions to contemporary Tunisian populations during prehistory and historic times have been diverse.
Today, Tunisia has a population of about 11 millions represented by Berbers, Arabs, Andalusians, Jews, Europeans and people of sub-Saharan origins. Tunisian cosmopolitan populations are situated on coastal locations. One of such city is Sousse, founded in the eleventh century B.C. as Hadrumetum by the Phoenicians it developed into an important center within the Carthaginian dominion. Sousse soon became the most important trading post on the North African coastline. Through history Hadrumetum came under the control of a number major cultures including the Vandals after the fall of the Roman Empire in the sixth century and later by the Byzantines that renamed it Hunerikopolis and Justinianopolis, respectively. The city became one of the most important Byzantine bases in North Africa. In the seventh century, the Arabs conquered the city renaming it Susa and introducing the Islamic religion and Arabic language.8 The city became a prosperous seaport during the Islamic Aghlabid Dynasty, which occupied and controlled Northern Africa for several centuries.
Subsequently, Sousse was invaded by the Normans of Sicily in the twelveth century9 followed by the Spanish. During the sixteenth century, Sousse received additional, but limited, contributions from the Ottoman Turks. Later, the city came under the control of the French, who once again renamed it to its current name of Sousse.10 Considering this complex history, the expectation is that the genetic landscape of Sousse has been shaped by varying degrees of influences from the above listed invaders who occupied the region in different periods of its history. In addition, Sousse is the most ancient settlement in Tunisia with uninterrupted habitation since its foundation by the Phoenicians.11 Towns such as Carthage and Utique, also established by the Phoenicians,12, 13 were destroyed and their population dispersed. Berber groups, on the other hand, have been rather isolated, since their towns and villages were not located on the coast.14 Studies on Tunisian cosmopolitan populations often examine Tunis which was founded later than Sousse and does not provide a prolong continuous antiquity. Hence, the case of the Sousse population represents an unique case study in population genetics. Despite its historical importance and strategic location, only one molecular genetics study has been performed on this anthropological interesting population.15
Over the past decade, the field of human population genetics has expanded our understanding of the origins, evolutionary history and migration patterns of human populations. Y-chromosome polymorphisms are ideally suited for studies of the patrilineal influences in highly admixed groups16, 17 such as the Sousse population of the present study residing in North Africa, a region characterized by a complex history of demographic events. Previous studies of North African populations have uncovered diverse genetic compositions18, 19, 20 as well as a high frequencies of two specific North African haplogroups (E-M81 and E-M78). Our previous investigation based on Y-chromosome polymorphisms21 using different Tunisian ethnic groups (Andalusian, Cosmopolitan Arab and three Berber-speaking groups) indicate high frequency of the autochthonous and most common North African haplogroup E-M81 (71%). The findings published by Ennafaa et al.22 also demonstrate that the E-M81 lineage is the most abundant (48.2%) in all Tunisian collections studied (Berbers from Bou Omran and Bou Saad located in southern Tunisia and Berbers and Arabs from the Jerba Island in South Tunisia). In general, all studies of North African populations18, 19, 20, 23 are in agreement in that the North African-specific lineage E-M81 is frequent in Berber-speaking groups.19, 20, 23 This observation suggests that the Middle Eastern influence during Neolithic and Arab domination had a greater cultural impact rather than a genetic influence in North Africa. In fact, the presence of the Near Eastern lineage J-M267, detected in the Tunisian samples at frequencies ranging from 17 to 30%,21, 22 is consistent with differential levels of paternal gene flow from the Near East that would have accompanied the Islamic invasion and previous dispersals from the Middle East during the Neolithic. In addition, a sub-Sahara African component is also detected in these populations as attested by haplogroups A, B, E-M96, E-M2 and E-M35 (16.3%) resulting, at least partially, from slavery or migration in recent times.
In spite of these studies, the current understanding of Y-chromosome variation in Tunisia is limited compared with what is known of mitochondrial DNA diversity. In addition, coastal regions, such as Sousse, have been underrepresented in most studies of Tunisian genetic variation. Previous studies based on Y-chromosome-single-nucleotide polymorphism (Y-SNP) data from Tunisia21, 22 have focused mainly on the Berber groups and the Andalusians. The present study represents the first one of its kind to examine the coastal population of Sousse involving a considerable sample size and comprehensive phylogenetic analyses.
In this study, we complement our previous Y-chromosome-short tandem repeat (Y-STR) data based on 17 Y-STR loci15 by performing, for the first time, a high-resolution analysis based on 51 Y-SNPs in the Sousse population. The aim of the current investigation is to elucidate the paternal genetic structure throughout Tunisia and provide information on the complex pattern of migration and admixture expected from historical data. Sousse, as the most ancient cosmopolitan region in Tunisia may reveal most of the genetic contributions that have influenced the present day Tunisian populace.
Materials and methods
Population studied
A total of 220 unrelated healthy males from the region of Sousse were examined employing Y-SNPs. The samples constitute a set of that typed for Y-STR,15 and therefore, the same selection criteria and DNA extraction procedure were employed.
Y-SNP genotyping
Fifty-one Y-SNPs markers were genotyped in a hierarchical manner using standard methods, including polymerase chain reaction-restriction length fragment polymorphism, the YAP polymorphic Alu insertion, allele-specific polymerase chain reaction (for primer sequences and polymerase chain reaction conditions see Supplementary Table S6) and direct sequencing.24 Initially, all the samples were analyzed for an insertion polymorphism, the YAP (a Y-specific polymorphic Alu insertion) or PAI, that define E lineages.
For the remaining SNPs, the following typing scheme was used:
-
Samples carrying the Alu insertion were genotyped for the biallelic markers M96, M2, M215, M329, M75, M35, M78, M81 and M123. Samples derived for M81 were tested for M107, M165 and M183, and individuals derived for M78 were tested for V12, M224, V13, V22 and V65.
-
Samples without Alu insertion were hierarchically screened for the SNPs M304, P58 and M410. Next, M367, M368, M369, L147.1, L174, L222.2, L65.2 were typed in samples carrying the P58 mutation and lacking M410. All the individuals derived for the marker M410 were tested for Page 55, M67, M530/L24, M158 and DYS445⩽7.
-
Afterwards, M69, M522, P207, M213, V88, M168, M216, M214, M91, M198, M42, M9, M258, M20, M201, M370, M269, M174 and M184 were screened in the remaining individuals, until all samples were assigned to a specific terminal subhaplogroup.
The phylogenetic relationship of these markers and the haplogroups that they define are indicated in Figure 1. The Y-SNP haplogroup nomenclature follows the recommendation originally proposed by Karafet et al.25 and revised by additional lineage information as established by subsequent scientific teams.26, 27
Statistical analyses
A total of 23 key geographically targeted reference populations previously typed for both Y-STR and Y-SNP markers and published were utilized for the phylogenetic analyses (Supplementary Table S1). They correspond to 17 countries in North Africa, the Near East and Europe as well as some sub-Saharan African countries (Supplementary Figure S1).
Frequency distribution maps for haplogroups (contour map) were obtained using the Surfer v8.07 software (Golden Software, Golden, CO, USA, http://www.goldensoftware.com). Previously published data on African and Arabian populations were included to build the grid (Supplementary Table S2). We employed the ordinary Kriging procedure with default settings for interpolating frequency values. Kriging variance was used to compute the confidence interval limits of the estimated values. Kriged values falling out of the confidence interval limits were not considered in the analyses. To prevent the Kriging interpolation from performing unrealistic gradient estimation, boundaries of large uninhabitable areas were defined. (See Supplementary Figure S1 for the location of the observed values). Phylogenetic relationships among the microsatellite haplotypes belonging to haplogroups E-M81, E-M78, J-M267, J-M172 and R-M207 were inferred through median joining networks using the NETWORK 4.6.0.0 (Fluxus Engineering, Suffolk, UK)28 and Network Publisher version 1.2.0.0 (Fluxus Engineering, www.fluxusengineering.com) softwares, using weighting based on the inverse of the microsatellite variances. The networks generated are based on the following 10 Y-STR loci: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439. These loci are in common among all relevant reference populations (Supplementary Table S2) included in the analyses. DYS385 a/b were excluded from the analyses since the multiplex amplification reactions do not allow for the discrimination between the two loci. DYS389II was scored by subtracting the DYS389I allele from the total fragment.29 Times to the most common recent ancestor for the most diagnostic Y haplogroups were estimated by calculating the mean STR variance as proposed by Kayser et al.30 using a mean STR mutation rate of 0.000 69 per generation of 25 years.31
Phylogenetic relationships
The genetic relationships among Sousse and the reference populations were analyzed with two approaches. The first approach was based on genetic distances (using Rst) calculated with the Arlequin program v3.5(University of Bern, Bern, Switzerland)32 and displayed in a Multidimensional Scaling (MDS) graph using the STATISTICA 8 package (Statistica, SAS/STAT Software, Cary, NC, USA; http://www.statsoft.com). Genetic distances were estimated at the level of haplotypes based on the ten aforementioned Y-STR loci (DYS19, DYS389I, DYS389b, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439). The second scheme was based on relative haplogroup frequencies normalized within populations, centered, and without variance normalization in order to construct a principal component analysis (PCA) plot. To study the population genetic structure in North African and, specifically, of Tunisian populations, we performed a series of hierarchical analyses of molecular variance pooling populations according to geographical, linguistic and ethnical criteria. Two sets of analyses of molecular variances, one based on Y-STR and the other on Y-SNP data, were performed using the Arlequin software ver. 3.5.32
Results
Y-chromosome lineage diversity
The partitioning of paternal lineages and their frequencies are displayed in a hierarchical phylogeny in Figure 1. The 220 Sousse Y-chromosomes represent 24 different haplogroups, the majority of them belonging to haplogroups E and J that account for 90% of our dataset (E; 56% and J; 34%) (Supplementary Table S3). The predominant E sublineage, also commonly found in other North African populations,18, 19, 20, 21, 22 is E-M81. All E-M81 derived chromosome are in subhaplogroup E-M183 (44.55%). Besides the common E-M81 lineage, traces of other lineages within the major E-M215 haplogroup were detected including E-M35, E-M78 and E-M123. These lineages are also found at various frequencies throughout North and East Africa. Haplogroup E-M78 which has a wide distribution, including Europe, the Near East and North Africa,20 is mostly represented by subhaplogroup E-V65 (4.09%), with the exception of two individuals that belong to subhaplogroups E-V13 and E-V22 (0.45% each). Lineage E-M123, on the other hand, is detected at low frequency (1.82%). It is interesting that the E-M2 clade which is particularly frequent in sub-Saharan Africa33, 34 and present in some North African populations most likely as a result of sub-Saharan migration18, 19, 35, is not detected in the Sousse samples. The second most prevalent major lineage detected in our dataset is J-M304 encompassing 34% of the total male population. Both of its main branches, J-P58 and J-M172, were observed. Haplogroup J-P58, which is thought to have originated in eastern Anatolia,36 is largely represented by subhaplogroup J-L-222.2 (23.64%). Within the J-M172 clade, the majority of males are represented by the J-L24 subclade (5.45%). Furthermore, Y-haplogroup R-M207 is found in 12 Sousse males of which three individuals are either R-M198 (0.45%) or R-V88 (0.91%). Also present were haplogroups G-M201, L-L20, T-M184 and A-M91, although each of these accounted for only 0.45% of the entire sample set.
High resolution contour maps based on frequency distributions of informative haplogroups were performed. The maps indicate distinct west–east genetic clines in North Africa. The frequency of haplogroup E-M81 (Figure 2a), for example, is much higher in northwestern Africa particularly in Tunisian and moderate in the Near East and Europe. On the other hand, the frequencies of haplogroups E-M78 and E-M123 are much higher in northeast Africa exhibiting a focal point of extreme frequencies in Egypt-Palestine (Figures 2b and c, respectively). The J-M267 lineage is prevalent in all North African and Levantine groups (Figure 2d). In North Africa, J-M267 exhibits the highest frequency in Andalusians from Zaghouan. It is also found at relatively high frequency in the Levantine samples. The J-P58 sublineage contour map is depicted in Figure 2e.
Phylogeographic analyses of prominent haplogroups
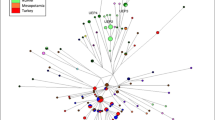
To examine Y-chromosomal relationships in more detail, networks analyses were performed utilizing the 10 Y-STR markers common to all populations. The projection illustrating Y-STR haplotype variation identified in 489 Y-chromosomes belonging to the E-M81 haplogroup is presented in Figure 3a. It is interesting to note that the E-M81 mutation was not detected in the Egyptian population analyzed in this study. The star-shaped E1b1b1b-M81 network is centered on the most frequent and widespread haplotype seen in all North African and European populations. In addition, several other shared haplotypes were observed, with the majority of these present in North African populations. The sharing of haplotypes suggests relatedness among the samples. On the other hand, a greater number (51) of North African-specific clades are also noted, further attesting to the high level of diversity observed throughout the region. It is noteworthy that of all Tunisian populations analyzed, Sousse possesses several unique haplotypes (13/27), indicating a high level of microsattelite diversity. The overall haplotype diversity (HD) and the phylogenetic Mean Pairwise Distance (MPD) values within haplogroup E-M81 are 0.8362±0.0151 and 2.1126±1.1799, respectively. The coalescence time estimate for the E-M81 network is 5.7±3.9 kya.
Median joining networks based on Y-chromosome-short tandem repeat haplotypes within haplogroups E-M81 (a), E-M78 (b), J-M267 (c), J-M172 (d) and R-207 (e) of North Africans, Middle Easterners, European and sub-Saharan populations. The circle sizes are proportional to the haplotype frequencies. The smallest area is equivalent to one individual. Branch lengths are proportional to the number of mutational steps separating two haplotypes. A full color version of this figure is available at the Journal of Human Genetics journal online.
The topology of the E-M78 network (Figure 3b) exhibits greater diversity compared with that of E-M81. The E-M78 haplogroup is mostly observed in the Near East populations. Noteworthy, with the exception of the Sousse population, this haplogroup was more frequent in northeastern (Libya; 12.7% and Egypt; 33%) compared with northwestern Africa. Also interesting, Sousse, in contrast to the other Tunisian populations, possesses the highest number of Y-STR- E-M78 haplotypes (18/13, respectively), three of them unique and the ten remaining shared among Europeans, Near Easterners and North Africans (Tunisian Andalusians from Zaghouan, Libyans, Moroccans and Egyptians). This haplotype sharing may be indicative of gene flow involving these regions. The diversity values within haplogroup E-M78 are higher than for E-M81 (0.9899±0.0017 and 4.1310±2.03985, for HD and MPD respectively). The age estimation for E1b1b1a-M78 is 12.2±9.2 kya.
The J-M267 network includes 500 haplotypes, most of them are from Near Easterners (Figure 3c). The most abundant haplotype consists of 106 individuals, including Libyans (42), Soussians (21) and Near Easterners (21). The 22 remaining haplotypes include people from Algeria, Morocco and the Tunisian ethnic groups. In this J-M267 projection, extensive haplotype sharing between the Sousse population and different North African and Near Eastern groups is observed as well as the presence of 10 haplotypes unique to Sousse males. Diversity estimates within haplogroup J-M267 are 0.9439±0.0073 and 2.8360±1.4977 for HD and MPD, respectively. The age estimation for J1-M267 is 7.6±5.2 kya.
The J-M172 network (Figure 3d) is mostly represented by Near Eastern samples (73.9%) with a moderate number of European individuals (18.5%) and only a few North African haplotypes (11.7%). Within North African populations, the Sousse population exhibits the highest number of haplotypes (18). A total of 41 haplotypes have been detected in North African populations. The J-M172 topology illustrates limited haplotype sharing, mostly between Near Easterners and Europeans. Furthermore, numerous Near Eastern- and European-specific haplotypes with highly diverse distribution were noted. Diversity estimates within haplogroup J-M172 are 0.9932±0.0011 and 4.6173±2.2714 for HD and MPD, respectively, and its age estimate is 15.8± 7.1 kya.
The network projection for individuals harboring the R-M207 mutation is provided in Figure 3e. Haplogroup R-M207 was frequent in European groups and less abundant in Near Easterns. This clade is sporadically detected in sub-Saharan and in North African populations. With regards to Tunisian collections, R-M207 is found only in Sousse and Andalusians samples. Diversity values within haplogroup R-M207 are 0.9933± 0.0013 and 4.8067±2.3515 for HD and MPD, respectively.
Genetic structure
To explore the genetic relationships among the populations of Sousse, Tunisia as a whole and the 23 other geographically targeted populations obtained from the literature (Supplementary Table S1), we performed a PCA based on haplogroup frequencies (Supplementary Table S2). The first two components of the PCA account for 55.18% of the variation and reveal distinct geographical partitioning (Figure 4a). The North African populations form a cluster located in the upper-left portion of the plot, except for the Egyptians that lie closest to Palestinians to the lower right. This cluster of North African populations is defined by the predominance of haplogroup E-M81. In contrast, Egyptians are characterized by high frequency of the E-M78 haplogroup and the absence of the E-M81 lineage. Within the North Africa cluster, the Sousse sample is close to the Cosmopolitans and Andalusians Tunisian groups and the general populations from neighboring states, namely Libya, Algeria and Morocco. The Tunisian Berber collections form a tide conglomerate isolated in the upper-left corner of the graph. The sub-Saharan Africans which possess higher frequencies of haplogroups B-M96, A-M91, E-M2 and E*-M96 lie close to each other in the lower left of the plot. The PCA also illustrate the genetic affinity of Levantine populations to Europeans especially Italians. In fact, both groups present relatively high frequencies of J-M172 and share some other lineages in particular I-M170 and T-M70.
(a) Principal component analysis of Y-chromosomal haplogroup frequency data from the investigated and reference populations. (b) Multidimensional scaling plot based on Rst genetic distances of the same populations used in the principal component analysis. The acronyms are the same as those used in Supplementary Table S1. A full color version of this figure is available at the Journal of Human Genetics journal online.
The populations were also contrasted using Rst estimates (Supplementary Table S4) computed from the 10 loci Y-STR haplotypes and plotted using multidimensional scaling (MDS) (Figure 4b). The MDS illustrate similar general features as the PCA, including the North Africans cluster close to Near Easterners with the Sousse population partitioning similarly relative to the other groups as well as the Egyptians affinity to the Palestinians. Yet in the MDS the Near Easterners and Tuareg segregate towards sub-Saharan Africans which, like in the PCA graph, aggregate in the lower-left corner of the plot distant from all other populations.
To further explore genetic structure in our samples, several separate analyses of molecular variances were performed using both Y-STR haplotypes and Y-SNP haplogroups data (Supplementary Table S5). A significant genetic heterogeneity was revealed when all populations were considered as a single group. When considering North Africa versus the three geographical regions separately (Near East, Europe and sub-Sahara Africa), the only non-significant genetic differentiation was relative to the Near East.
Discussion
This study characterizes the Y-chromosome DNA diversity of the Sousse population using high-resolution biallelic and STR markers coupled to a wide sampling coverage of reference populations in order to provide a comprehensive understanding of the paternal genetic substructure of the Tunisian people and their origins in the content of the North African genetic landscape. Our results indicate that the frequency of the E-M81 haplogroup in the region of Sousse is relatively lower than in Berber group; it reaches a value of 45%, comparable with the values detected in the Cosmopolitan population from Tunis and Andalusians.21 This frequency range of 36–45% of E-M81 in these Cosmopolitan and Andalusian Tunisian populations is consistent with a strong common Berber back-ground that gives the typical profile to North African populations that aggregate in the same clusters in the PC and MDS analyses. However, depending on the Berber or Cosmopolitan status, North African populations can be classified into subclusters. This observation is in agreement with the analyses of molecular variance analyses that indicate significant differences in molecular variance for both SNP and STR marker systems (17.65 P⩽0.05 and 1.64 P⩽0.05, respectively) between Cosmopolitans and Berbers. In all of these parameters, the Sousse population exhibits characteristics of a highly genetically diverse North African population. Among the various Tunisian communities, haplogroup E-M81 is more prevalent in Berbers,21, 22 especially those inhabiting remote villages where it was found to be fixed in two independent Berber populations (Chenini-Douiret and Jradou).21 Similarly, previous phylogeographic analyses of the E-M81 lineage in other North African Berber-speaking groups has shown its high frequency (80% in the Mozabites of Algeria; 65%–73% in Berbers from Morocco19, 20, 23).
The E-M81 lineage exhibits a star-like network structure (Figure 3a), which suggests an ancient evolution. This network exhibits apparent rapid expansion at some point that may be explained by loss of diversity due to genetic drift. Indeed, most of the STR haplotypes belonging to the E-M81 haplogroup are shared among various North African communities without obvious genetic structure relative to geography. Based on these phylogeographic considerations, the E-M81 haplogroup is likely to represent an important paternal founder lineage of North African people.
In our previous work performed on Tunisian populations, this haplogroup was dated to 7.4±5.5 kya in the Neolithic, comparable with the age estimated in this study (5.7±3.9 kya), and by Arredi and collaborators.19 However, considering the high level of genetic drift typically experienced by uniparental marker systems such as the Y-chromosome, it is possible that the E-M81 haplogroup had a more ancient genesis in North Africa. Indeed, according to Keita,37 E-M81 most likely emerged from the sub-Saharan clade E-M35 either in the Maghreb region, or possibly as far south as the Horn of Africa. This conjecture is consistent with the creation of sub-Saharan mitochondrial DNA markers that date to 20 kya38 and probably contemporary with the Ibero-Maurusian civilization during the Paleolithic.
Along with the high prevalence of E-M81 in Sousse, we observed the E-M78 mutation at low frequency (5%). In contrast to the above-mentioned E-M81 haplogroup, E-M78 has a wide geographic distribution with its highest frequency observed in the Egypto-Palestinian area. It is detected at lower frequencies in Northwest Africa and is particularly observed in Andalusians and Cosmopolitans rather than Berbers. This suggests an east to west gene flow with greater penetration into the Cosmopolitan populations of North Africa.
In addition to the indigenous components, gene flow from the Middle East is also evident in certain Tunisian populations by the presence of the J-M304 haplogroup. In fact, J-M304 is the predominant Y-chromosome lineage in the Middle East. This lineage splits into two main subclades, J-M267 and J-M172. The frequency and diversity of J-M267 is highest in the Middle East. In Tunisia, with the exception of three Berber communities, J-M267 is detected with frequencies ranging from 48% to 5%. In Sousse J-M267 is observed at levels (26%) comparable with that found in Cosmopolitans from Tunis (28%). The network topology of the J-M267 lineage illustrates a star-like topology with no geographic structuring. This feature supports a demic expansion from ancestral haplotypes currently shared by Maghreban and Middle Eastern populations, subsequent to migrations from the east. J-M267 and its subhaplogroup J-P58 are widely distributed in North Africa but display higher frequencies in the east with the exception of some Tunisian regions.
Gene flow from the Middle East should be considered according to different migration waves during prehistory, antiquity and in more recent times. In prehistorical and historical times, Middle Eastern contributions to North African populations could have happened during Neolithic period around 8000 BP as part of the Capsian civilization developments that were introduced in North Africa along with agriculture. The presence of the J-M267 haplogroup with a long local evolution in the Berber of Sened in the nominal Capsian region is congruent with this hypothesis. However, its presence in the Sousse population at a relative high frequency may be indicative of additional secondary regional gene flow events.
Undoubtedly, the Muslim expansion of the seventh century into North Africa and the subsequent massive Bedouin migration during the eleventh century contributed considerably to the east to west gene flow into North Africa. The J-M267 haplogroup with frequencies as high as 40% in Saudi Arabia39 could have been introduced with the Arab migration waves into North Africa during historical times. Its presence in North Africa at frequencies around 30%21, 35, 40 and its absence in Berbers, excluding the Sened (see above), may be explained by the Arab conquest. Hence, it could be considered as an indicator of Arab gene flow into these populations. Furthermore, the relative high frequency of the J-M267 haplogroup in Andalusians from Tunisia may be the result of the strong relationship between Andalusia in southern Spain and Northwest Africa as well as the Middle East during the Omeyad dynasty, and the subsequent expatriation of Muslims from Iberia.
The Sousse population and the other Tunisian groups already studied exhibit a fair amount of differences in terms of their Middle Eastern paternal lineages composition. Sousse, particularly, possesses a relatively high frequency of haplogroup J-M172 (~9%) that is absent in all of the remaining Tunisian populations with the exception of Andalusians (~3%). Haplogroup J-M172 has been associated with population movements in the Fertile Crescent during the Neolithic Agricultural revolution. Today, it is very frequent in the Levant, Anatolia and Iran41, 42 and its recent spread in the Mediterranean is believed to have been facilitated by the maritime trading culture of the Phoenicians (1550–300 BC). According to Zalloua and collaborators43 evidence of Phoenician influence in Tunisian is apparent by the presence of the J-M172 Y-chromosome haplogroup in coastal regions considered as areas of Phoenician contact (versus inland). In Sousse, the J-M172 lineage is exclusively represented by its J-M410 clade, of which the J-L24 mutation is the most prevalent (12 individuals). The remaining seven individuals belong to the following subclades: J-M410 (three individuals), J-Page55 (two individuals) and J-DYS445⩽7 (one individual). In fact, J-L24, the most frequent subclade, constitutes also the most widespread subclade of J2a, with a geographic distribution ranging from the Middle East to Europe, North Africa and South Asia.
The J-M172 haplogroup associated with the Phoenician expansion is distributed throughout the Mediterranean basin and Asia. It is thought that the Phoenicians originated in what is today coastal Lebanon and subsequently founded and settled several city-states in the Mediterranean including in North Africa. In Tunisia, their population number was estimated, at the end of their dominion, to be 100 000 compared with 500 000 Berbers.44 It is important to note that although the most famous city founded by the Phoenicians was Carthage, they also established the settlements of Utique and Sousse.13 Interestingly, Sousse is the only Phoenician town in Tunisia that has been continuously inhabited since its foundation8 and it is the only population where the J-M172 Phoenician paternal marker is detected. In Sousse, J-M172 dates to 6.9±5.8 kya. These results are consistent with the J-M172 introduction into the Sousse population as a diversified haplotype and subsequently maintained locally. This interpretation is congruent with the Network analysis of this haplogroup. The J-M172 haplogroup is also observed in Andalusians. Andalusians are expected to have a direct Middle Eastern contribution in the Iberian Peninsula45 before their expatriation to North Africa after the fall of Granada to Christian forcces in 1492 A.D.
A more recent potential Middle Eastern genetic contribution to the North Africa gene pool may be associated with the expansion of the Ottoman Empire. Sousse also may have been specifically impacted by the Turkish occupation of North Africa. Yet, the unique presence of J-M172 in Sousse and its absence from other Tunisian regions that were under Ottoman influence argues for J-M172 in Sousse as a Phoenician signal. Further, Sousse exhibits another haplogroup, T-M184, that is not detected in any other North African population attesting again to the Phoenicians contribution to that population. Haplogroup T-M184 is more common today in East Africa and it is thought to signal the spread of agriculture from the Fertile Crescent. Indeed, the oldest subclades and the greatest diversity of T-M184 are found in the Middle East, especially within the Fertile Crescent. Yet, T-M184 could also have been dispersed throughout the Mediterranean basin by the Phoenicians (1200–800 before the common era).
In this study, we have also detected other haplogroups in Sousse including R-M207, G-M201, L-M20 and A-M91 that have not been reported in other Tunisian communities attesting to the genetic heterogeneity of this region. Haplogroup R is mainly represented by R-M207 in Sousse (nine individuals) and its distribution includes all continents being fairly common in Europe, South Asia and Central Asia.46, 47 It also occurs in the Caucasus, Middle East and in some parts of sub-Sahara Africa.41, 48 In addition, its two subclades, R1a and R1b, are observed at low frequencies (0.45 and 0.91%, respectively) in Sousse. R1a is most prevalent in East Europe while R1b is most frequent among West Europeans.49
Although at very-low frequency (0.45%), it must be noted that haplogroupe A-M91, the oldest human haplogroup, is found in Sousse but nowhere else in North Africa. This haplogroup is mainly found in Eastern and Southern Africa. It is common among the Khoisan people, including the Bushmen, and it is considered an original ancestral haplogroup.
In conclusion, the analysis of admixed populations represents an unique opportunity to examine the impact of multiple migrations into a region. Within a historical context, both population isolation and admixture have had a considerable impact on the Tunisian population structure. The wide range of paternal lineages present in our Sousse population indicates a diverse origin. Indeed the genetic structure observed of paternal lineages in Sousse is more diversified than any other studied Tunisian population and reflects largely the influence of successive migrations since its foundation by the Phoenicians.
References
Séthom, H . & Kassab, A . Les régions géographiques de la Tunisie (University of Tunis, Tunisia, 1981).
Despois, J . Une géographie de la Tunisie (Editions Persée, Paris, France, 1969).
Boularès, H . Histoire de la Tunisie: Les grandes dates, de la Préhistoire à la Révolution, Cérès Edition (Tunis, Tunisia, 2012).
Camps, G . Les civilisations préhistoriques de l'Afrique du Nord et du Sahara, (Doin, Paris, 1974).
Camps, G . (ed.) Beginnings of pastoralism and cultivation in north-west Africa and the Sahara: origins of the Berbers, (Cambridge University Press, Cambridge, 1982).
Murdock, G. P . Africa, its peoples and their culture history, (McGraw-Hill Book Company, New York, Toronto, London, 1959).
Hiernaux, J . The people of Africa, (Charles Scribner's Sons, New York, 1975).
Djelloul, N . Sousse ancient Hadrumetum Contraste Edn (Oloron-Sainte-Marie, France, 2006).
Norwich, J. J . The Normans in Sicily (Penguin Global, London, UK, 1992).
Pétridès, S . ‘Hadrumetum’ the Catholic encyclopedia, (Robert Appleton Company, New York, 1910).
said, B. e. n . & Tunisian, F . Coastal cities attractiveness and amenities MPRA Paper No. 52961 (2014).
Lancel, S . Carthage, (Edition PUF, Paris, 1982).
Fantar, M. H . Carthage, Approche d’une civilisation, (Edition Alif, Tunis, 1993).
Ibn Khaldoun, A . Histoire des Berbères et des dynasties musulmanes de l’Afrique septentrionale. Traduction de Le Baronde Slane, (Paul Geuthner, Paris, 1968).
Fadhlaoui-Zid, K ., Chennakrishnaiah, S ., Zemni, R ., Grinberg, S ., Herrera, R. J . & Benammar-Elgaaied, A . Sousse, Tunisia: tumultuous history and high Y-STR diversity. Electrophoresis 33, 3555–3563 (2012).
González-Andrade, F ., Sánchez, D ., González-Solórzano, J ., Gascón, S . & Martínez-Jarreta, B . Sex-specific genetic admixture of Mestizos, Amerindian Kichwas, and Afro-Ecuadorans from Ecuador. Hum. Biol. 79, 51–77 (2007).
Mendizabal, I ., Sandoval, K ., Berniell-Lee, G ., Calafell, F ., Salas, A . & Martínez-Fuentes, A . et al. Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba. BMC Evol. Biol. 21, 213 (2008).
Bosch, E ., Calafell, F ., Comas, D ., Oefner, P. J ., Underhill, P. A . & Bertranpetit, J . High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian peninsula. Am. J. Hum. Genet. 68, 1019–1029 (2001).
Arredi, B ., Poloni, E. S ., Paracchini, S ., Zerjal, T ., Fathallah, D. M . & Makrelouf, M . et al. A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa. Am. J. Hum. Genet. 75, 338–345 (2004).
Cruciani, F ., La Fratta, R ., Santolamazza, P ., Sellitto, D ., Pascone, R . & Moral, P . et al. Phylogeographic analysis of haplogroup E3b (E-M215) y chromosomes reveals multiple migratory events within and out of Africa. Am. J. Hum. Genet. 74, 1014–1022 (2004).
Fadhlaoui-Zid, K ., Martinez-Cruz, B ., Khodjet-el-khil, H ., Mendizabal, I ., Benammar-Elgaaied, A . & Comas, D . Genetic structure of Tunisian ethnic groups revealed by paternal lineages. Am. J. Phys. Anthropol. 146, 271–280 (2011a).
Ennafaa, H ., Fregel, R ., Khodjet-El-Khil, H ., Gonzalez, A. M ., Mahmoudi, H. A . & Cabrera, V. M . et al. Mitochondrial DNA and Y-chromosome microstructure in Tunisia. J. Hum. Genet. 56, 734–741 (2011).
Semino, O ., Magri, C ., Benuzzi, G ., Lin, A. A ., Al-Zahery, N . & Battaglia, V . et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 74, 1023–1034 (2004).
Gayden, T ., Regueiro, M ., Martinez, L ., Cadenas, A. M . & Herrera, R. J . Human Y-chromosome haplotyping by allele-specific polymerase chain reaction. Electrophoresis 29, 2419–2423 (2008).
Karafet, T. M ., Mendez, F. L ., Meilerman, M. B ., Underhill, P. A ., Zegura, S. L . & Hammer, M. F . New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 18, 830–838 (2008).
Underhill, P. A ., Myres, N. M ., Rootsi, S ., Metspalu, M ., Zhivotovsky, L. A . & King, R. J . et al. Separating the post-glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur. J. Hum. Genet. 18, 479–484 (2010).
Myres, N. M ., Rootsi, S ., Lin, A. A ., Jarve, M ., King, R. J . & Kutuev, I . et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 19, 95–101 (2011).
Bandelt, H. J ., Forster, P . & Rohl, A . Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Rolf, B ., Meyer, E ., Brinkmann, B . & de knijff, P . Polymorphism at the tetranucleotide repeat locus DYS389 in 10 populations reveals strong geographic clustering Eur. J. Hum. Genet. 6, 583–588 (1998).
Kayser, M ., Krawczak, M ., Excoffier, L ., Dieltjes, P ., Corach, D . & Pascali, V . et al. An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am. J. Hum. Genet. 68, 990–1018 (2001).
Zhivotovsky, L. A ., Underhill, P. A ., Cinnioglu, C ., Kayser, M ., Morar, B . & Kivisild, T . et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 74, 50–61 (2004).
Excoffier, L . & Lischer, H. E . Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Underhill, P. A ., Shen, P ., Lin, A. A ., Jin, L ., Passarini, G . & Yang, W. H . et al. Y chromosome sequences variation and the history of human populations. Nat. Genet. 26, 358–361 (2000).
Cruciani, F ., Santolamazza, P ., Shen, P. D ., Macaulay, V ., Moral, P . & Olckers, A . et al. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am. J. Hum. Genet. 70, 1197–1214 (2002).
Robino, C ., Crobu, F ., Di Gaetano, C ., Bekada, A ., Benhamamouch, S . & Cerutti, N . et al. Analysis of Y chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample. Int. J. Legal Med. 122, 251–255 (2008).
Chiaroni, J ., King, R. J ., Myres, N. M ., Henn, B. M ., Ducourneau, A . & Mitchell, M. J . et al. The emergence of Y-chromosome haplogroup J1e among Arabic-speaking populations. Eur. J. Hum. Genet. 18, 348–353 (2010).
Keita, S . Geography, selected Afro-Asiatic families, and Y chromosome lineage variation: an exploration in linguistics and phylogeography In: Bengtson JD editor. In hot pursuit of language in prehistory, John Benjamins Publishing Company: Amsterdam, 3–16 (2008).
Frigi, S ., Cherni, L ., Fadhlaoui-zid, K . & Benammar-Elgaaied, A . Ancient local evolution of African mtDNA haplogroups in Tunisian Berber populations. Hum. Biol. 82, 367–384 (2010).
Abu-Amero, K ., Hellani, A ., González, A ., Larruga, J. M ., Vicente, M. C . & Underhill, P . Saudi Arabian Y-chromosome diversity and its relationship with nearby regions. BMC Genet. 59, 1–9 (2009).
Fadhlaoui-Zid, K ., Haber, M ., Martínez-Cruz, B ., Zalloua, P ., Benammar-Elgaaied, A . & Comas, D . Genome-wide and paternal diversity reveal a recent origin of human populations in North Africa. PLoS ONE 8, e80293 (2013).
Zalloua, P. A ., Xue, Y ., Khalife, J ., Makhoul, N ., Debiane, L . & Platt, D. E . et al. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am. J. Hum. Genet. 82, 873–882 (2008a).
El-Sibai, M ., Platt, D. E ., Haber, M ., Xue, Y ., Youhanna, S. C . & Wells, R. S . et al. Geographical structure of the Y-chromosomal genetic landscape of the Levant: a coastal-inland contrast. Ann. Hum. Genet. 73, 568–581 (2009).
Zalloua, P. A ., Platt, D. E ., El Sibai, M ., Khalife, J ., Makhoul, N . & Haber, M . et al. Identifying genetic traces of historical expansions: Phoenician footprints in the Mediterranean. Am. J. Hum. Genet. 83, 633–642 (2008b).
Julien, C. A . Histoire de l’Afrique du Nord, (Payot, Paris, 1961).
Abdul-Wahab, H. H . Coup d’oeil général sur les apports ethniques étrangers en Tunisie. Revue Tunisienne 24, 305–316 (1917).
Adams, S ., Bosch, E ., Balaresque, P. L ., Ballereau, S. J ., Lee, A. C . & Arroyo, E . et al. The genetic legacy of religious diversity and intolerance: paternal lineages of Christians. Jews, and Muslims in the Iberian Peninsula. Am. J. Hum. Genet. 83, 725–736 (2008).
Sengupta, S ., Zhivotovsky, L ., King, R ., Mehdi, S ., Edmonds, C . & Chow, C . et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am. J. Hum. Genet. 78, 202–221 (2006).
Montano, V ., Ferri, G ., Marcari, V ., Batini, C ., Anyaele, O . & Destro-Bisol, G . et al. The Bantu expansion revisited: a new analysis of Y chromosome variation in Central Western Africa. Mol. Ecol. 20, 2693–2708 (2011).
Myres, N. M ., Rootsi, S ., Lin, A. A ., Järve, M ., King, R. J . & Kutuev, I . et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 19, 95–101 (2011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Fadhlaoui-Zid, K., Garcia-Bertrand, R., Alfonso-Sánchez, M. et al. Sousse: extreme genetic heterogeneity in North Africa. J Hum Genet 60, 41–49 (2015). https://doi.org/10.1038/jhg.2014.99
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2014.99
This article is cited by
-
Insights into the Middle Eastern paternal genetic pool in Tunisia: high prevalence of T-M70 haplogroup in an Arab population
Scientific Reports (2021)
-
Human glutathione peroxidase codon 198 variant increases nasopharyngeal carcinoma risk and progression
European Archives of Oto-Rhino-Laryngology (2021)
-
Genetic variation of 17 X-chromosome STR loci in Tunisian population of Nabeul
International Journal of Legal Medicine (2019)
-
Contribution of Nitric oxide synthase 3 genetic variants to nasopharyngeal carcinoma risk and progression in a Tunisian population
European Archives of Oto-Rhino-Laryngology (2019)