Abstract
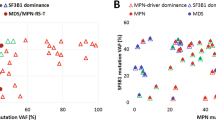
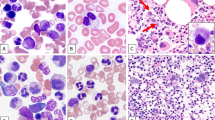
Chronic myeloid malignancies are categorized to the three main categories myeloproliferative neoplasms (MPNs), myelodysplastic syndromes (MDSs) and MDS/MPN overlap. So far, no specific genetic alteration profiles have been identified in the MDS/MPN overlap category. Recent studies identified mutations in SET-binding protein 1 (SETBP1) as novel marker in myeloid malignancies, especially in atypical chronic myeloid leukemia (aCML) and related diseases. We analyzed SETBP1 in 1 130 patients with MPN and MDS/MPN overlap and found mutation frequencies of 3.8% and 9.4%, respectively. In particular, there was a high frequency of SETBP1 mutation in aCML (19/60; 31.7%) and MDS/MPN unclassifiable (MDS/MPN, U; 20/240; 9.3%). SETBP1 mutated (SETBP1mut) patients showed significantly higher white blood cell counts and lower platelet counts and hemoglobin levels than SETBP1 wild-type patients. Cytomorphologic evaluation revealed a more dysplastic phenotype in SETBP1mut cases as compared with wild-type cases. We confirm a significant association of SETBP1mut with −7 and isochromosome i(17)(q10). Moreover, SETBP1mut were strongly associated with ASXL1 and CBL mutations (P<0.001 for both) and were mutually exclusive of JAK2 and TET2 mutations. In conclusion, SETBP1mut add an important new diagnostic marker for MDS/MPN and in particular for aCML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 4th edn. International Agency for Research on Cancer (IARC): Lyon, 2008.
Vardiman JW, Bennett JM, Bain B, Brunning RD, Thiele J . Atypical chronic myeloid leukaemia, BCR-ABL1 negative. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. (eds) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 4th edn. International Agency for Research on Cancer (IARC): Lyon, p 80–81 2008.
Fend F, Horn T, Koch I, Vela T, Orazi A . Atypical chronic myeloid leukemia as defined in the WHO classification is a JAK2 V617F negative neoplasm. Leuk Res 2008; 32: 1931–1935.
Bacher U, Schnittger S, Kern W, Weiss T, Haferlach T, Haferlach C . Distribution of cytogenetic abnormalities in myelodysplastic syndromes, Philadelphia negative myeloproliferative neoplasms, and the overlap MDS/MPN category. Ann Hematol 2009; 88: 1207–1213.
Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood 2005; 106: 2162–2168.
Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005; 7: 387–397.
Tefferi A, Gilliland DG . JAK2 in myeloproliferative disorders is not just another kinase. Cell Cycle 2005; 4: 1053–1056.
Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med 2007; 356: 459–468.
Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med 2006; 3: e270.
Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M et al. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood 2006; 108: 3472–3476.
Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol 2010; 28: 3858–3865.
Gelsi-Boyer V, Trouplin V, Adelaide J, Bonansea J, Cervera N, Carbuccia N et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol 2009; 145: 788–800.
Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood 2012; 120: 3080–3088.
Brecqueville M, Rey J, Bertucci F, Coppin E, Finetti P, Carbuccia N et al. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer 2012; 51: 743–755.
Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet 2013; 45: 18–24.
Hoischen A, van Bon BW, Gilissen C, Arts P, van LB, Steehouwer M et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet 2010; 42: 483–485.
Cristobal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood 2010; 115: 615–625.
Albano F, Anelli L, Zagaria A, Coccaro N, Casieri P, Minervini A et al. SETBP1 and miR_4319 dysregulation in primary myelofibrosis progression to acute myeloid leukemia. J Hematol Oncol 2012; 5: 48.
Panagopoulos I, Kerndrup G, Carlsen N, Strombeck B, Isaksson M, Johansson B . Fusion of NUP98 and the SET binding protein 1 (SETBP1) gene in a paediatric acute T cell lymphoblastic leukaemia with t(11;18)(p15;q12). Br J Haematol 2007; 136: 294–296.
Shaffer LG, Tommerup N . ISCN 2013: An international System for Human Cytogenetic Nomenclature. Karger: Basel, New York, 2013.
Haferlach C, Rieder H, Lillington DM, Dastugue N, Hagemeijer A, Harbott J et al. Proposals for standardized protocols for cytogenetic analyses of acute leukemias, chronic lymphocytic leukemia, chronic myeloid leukemia, chronic myeloproliferative disorders, and myelodysplastic syndromes. Genes Chromosomes Cancer 2007; 46: 494–499.
Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002; 100: 59–66.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Kumar P, Henikoff S, Ng PC . Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D . MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576.
Schnittger S, Bacher U, Kern W, Schroder M, Haferlach T, Schoch C . Report on two novel nucleotide exchanges in the JAK2 pseudokinase domain: D620E and E627E. Leukemia 2006; 20: 2195–2197.
Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood 2009; 113: 6182–6192.
Schnittger S, Eder C, Jeromin S, Alpermann T, Fasan A, Grossmann V et al. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia 2013; 27: 82–91.
Schnittger S, Bacher U, Haferlach C, Geer T, Muller P, Mittermuller J et al. Detection of JAK2 exon 12 mutations in 15 patients with JAK2V617F negative polycythemia vera. Haematologica 2009; 94: 414–418.
Schnittger S, Bacher U, Haferlach C, Dengler R, Krober A, Kern W et al. Detection of an MPLW515 mutation in a case with features of both essential thrombocythemia and refractory anemia with ringed sideroblasts and thrombocytosis. Leukemia 2008; 22: 453–455.
Grossmann V, Schnittger S, Kohlmann A, Eder C, Roller A, Dicker F et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood 2012; 120: 2963–2972.
Thiele J, Kwasnicka H, Orazi A, Tefferi A, Birgegard G . Polycythemia vera. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al. (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 4th edn. International Agency for Research on Cancer (IARC): Lyon, p 40–43 2008.
Tefferi A . Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia 2010; 24: 1128–1138.
Jeromin S, Haferlach T, Grossmann V, Alpermann T, Kowarsch A, Haferlach C et al. High frequencies of SF3B1 and JAK2 mutations in refractory anemia with ring sideroblasts associated with marked thrombocytosis strengthen the assignment to the category of myelodysplastic/myeloproliferative neoplasms. Haematologica 2012; 98: e15–e17.
Damm F, Itzykson R, Kosmider O, Droin N, Renneville A, Chesnais V et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia 2013; e-pub ahead of print 5 February 2013; doi:10.1038/lieu.2013.35.
Kanagal-Shamanna R, Bueso-Ramos CE, Barkoh B, Lu G, Wang S, Garcia-Manero G et al. Myeloid neoplasms with isolated isochromosome 17q represent a clinicopathologic entity associated with myelodysplastic/myeloproliferative features, a high risk of leukemic transformation, and wild-type TP53. Cancer 2012; 118: 2879–2888.
Schwaab J, Ernst T, Erben P, Rinke J, Schnittger S, Strobel P et al. Activating CBL mutations are associated with a distinct MDS/MPN phenotype. Ann Hematol 2012; 91: 1713–1720.
Schnittger S, Bacher U, Alpermann T, Reiter A, Ulke M, Dicker F et al. Use of CBL exon 8 and 9 mutations in diagnosis of myeloproliferative neoplasms and myelodysplastic/myeloproliferative disorders: an analysis of 636 cases. Haematologica 2012; 97: 1890–1894.
Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 2009; 460: 904–908.
Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 2011; 118: 3932–3941.
Gelsi-Boyer V, Trouplin V, Roquain J, Adelaide J, Carbuccia N, Esterni B et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol 2010; 151: 365–375.
Butler JS, Dent SY . The role of chromatin modifiers in normal and malignant hematopoiesis. Blood 2013; 121: 3076–3084.
Abdel-Wahab O, Adli M, Saunders L, Gao JG, Shih A, Pandey S et al. ASXL1 mutations promote myeloid transformation through inhibition of PRC2-mediated gene repression. Blood 2011; 118: 405a.
Oakley K, Han Y, Vishwakarma BA, Chu S, Bhatia R, Gudmundsson KO et al. Setbp1 promotes the self-renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10. Blood 2012; 119: 6099–6108.
Makishima H, Yoshida K, Nguyen N, Sanada M, Okuno Y, Ng KP et al. Somatic mutations in Schinzel-Giedion syndrome gene SETBP1 determine progression in myeloid malignancies. ASH Annual Meeting Abstracts 2012; 120: 2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
TH, WK, CH and SS are part owners of the MLL Munich Leukemia Laboratory GmbH. MM, UB and TA are employed by the MLL Munich Leukemia Laboratory GmbH. CG-P declares no conflict of interest.
Additional information
Author contributions
MM investigated molecular mutations, analyzed the data and wrote the manuscript; UB contributed to cytomorphologic analysis and classification of cases and wrote the manuscript; TA collected and documented clinical data and compiled statistical analyses; CG-P originally detected SETBP1 gene mutations and discussed with us this study; WK was involved in statistical analyses; CH was responsible for cytogenetics; TH was responsible for cytomorphologic analysis and was involved in the collection of clinical data; SS was the principal investigator of the study and wrote the manuscript. All authors read and contributed to the final version of the manuscript.
Rights and permissions
About this article
Cite this article
Meggendorfer, M., Bacher, U., Alpermann, T. et al. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia 27, 1852–1860 (2013). https://doi.org/10.1038/leu.2013.133
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2013.133
Keywords
This article is cited by
-
Identification of novel PIEZO1::CBFA2T3 and INO80C::SETBP1 fusion genes in an acute myeloid leukemia patient by RNA-seq
Molecular Biology Reports (2023)
-
Myelodysplastic/myeloproliferative neoplasms-unclassifiable with isolated isochromosome 17q represents a distinct clinico-biologic subset: a multi-institutional collaborative study from the Bone Marrow Pathology Group
Modern Pathology (2022)
-
A 19-year-old patient with atypical chronic myeloid leukemia
Annals of Hematology (2020)
-
The role of ASXL1 in hematopoiesis and myeloid malignancies
Cellular and Molecular Life Sciences (2019)
-
Diagnostik und Management der myelodysplastischen Syndrome*
InFo Hämatologie + Onkologie (2019)