Abstract
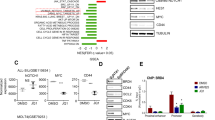
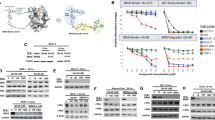
The PROTAC (proteolysis-targeting chimera) ARV-825 recruits bromodomain and extraterminal (BET) proteins to the E3 ubiquitin ligase cereblon, leading to degradation of BET proteins, including BRD4. Although the BET-protein inhibitor (BETi) OTX015 caused accumulation of BRD4, treatment with equimolar concentrations of ARV-825 caused sustained and profound depletion (>90%) of BRD4 and induced significantly more apoptosis in cultured and patient-derived (PD) CD34+ post-MPN sAML cells, while relatively sparing the CD34+ normal hematopoietic progenitor cells. RNA-Seq, Reverse Phase Protein Array and mass cytometry ‘CyTOF’ analyses demonstrated that ARV-825 caused greater perturbations in messenger RNA (mRNA) and protein expressions than OTX015 in sAML cells. Specifically, compared with OTX015, ARV-825 treatment caused more robust and sustained depletion of c-Myc, CDK4/6, JAK2, p-STAT3/5, PIM1 and Bcl-xL, while increasing the levels of p21 and p27. Compared with OTX015, PROTAC ARV-771 treatment caused greater reduction in leukemia burden and further improved survival of NSG mice engrafted with luciferase-expressing HEL92.1.7 cells. Co-treatment with ARV-825 and JAK inhibitor ruxolitinib was synergistically lethal against established and PD CD34+ sAML cells. Notably, ARV-825 induced high levels of apoptosis in the in vitro generated ruxolitinib-persister or ruxolitinib-resistant sAML cells. These findings strongly support the in vivo testing of the BRD4-PROTAC based combinations against post-MPN sAML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA . New mutations and pathogenesis of myeloproliferative neoplasms. Blood 2011; 118: 1723–1735.
Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood 2014; 123: e123–e133.
Rampal R, Mascarenhas J . Pathogenesis and management of acute myeloid leukemia that has evolved from a myeloproliferative neoplasm. Curr Opin Hematol 2014; 21: 65–71.
Keohane C, Mesa R, Harrison C . The role of JAK1/2 inhibitors in the treatment of chronic myeloproliferative neoplasms. Am Soc Clin Oncol Educ Book 2013, 301–305.
Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med 2010; 363: 1117–1127.
Vannucchi AM, Kantarjian HM, Kiladjian JJ, Gotlib J, Cervantes F, Mesa RA et al. A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis. Haematologica 2015; 100: 1139–1145.
Kundranda MN, Tibes R, Mesa RA . Transformation of a chronic myeloproliferative neoplasm to acute myelogenous leukemia: does anything work? Curr Hematol Malig Rep 2012; 7: 78–86.
Eghtedar A, Verstovsek S, Estrov Z, Burger J, Cortes J, Bivins C et al. Phase 2 study of the JAK kinase inhibitor ruxolitinib in patients with refractory leukemias, including post-myeloproliferative neoplasm acute myeloid leukemia. Blood 2012; 119: 4614–4618.
Fiskus W, Verstovsek S, Manshouri T, Rao R, Balusu R, Venkannagari S et al. Heat shock protein 90 inhibitor is synergistic with JAK2 inhibitor and overcomes resistance to JAK2-TKI in human myeloproliferative neoplasm cells. Clin Cancer Res 2011; 17: 7347–7358.
Meyer SC, Levine RL . Molecular pathways: molecular basis for sensitivity and resistance to JAK kinase inhibitors. Clin Cancer Res 2014; 20: 2051–2059.
Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 2012; 489: 155–159.
Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 2012; 119: 4480–4485.
Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci USA 2014; 111: E5401–E5410.
Belkina AC, Denis GV . BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 2012; 12: 465–477.
Shi J, Vakoc CR . The mechanism behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014; 54: 728–736.
Itzen F, Greifenberg AK, Bosken CA, Geyer M . Brd4 activates P-TEFB for RNA polymerase II CTD phosphorylation. Nucleic Acids Res 2014; 42: 7577–7590.
Nechaev S, Adelman K . Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 2011; 1809: 34–45.
Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR . Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334.
Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR . BET Bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell 2015; 58: 1028–1039.
Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI et al. Convergence of developmental and oncogenic signaling pathways at transcriptional superenhancers. Mol Cell 2015; 58: 362–370.
Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011; 478: 524–528.
Filippakapoulos P, Knapp S . Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014; 13: 337–356.
Boi M, Gaudio E, Bonetti P, Kwee I, Bernasconi E, Tarantelli C et al. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin Cancer Res 2015; 21: 1628–1638.
Basheer F, Huntly BJ . BET bromodomain inhibitors in leukemia. Exp Hematol 2015; 43: 718–731.
Saenz DT, Fiskus W, Manshouri T, Rajapakshe K, Krieger S, Sun B et al. BET protein bromodomain inhibitor-based combinations are highly active against post-myeloproliferative neoplasm secondary AML cells. Leukemia 2016; e-pub ahead of print 25 October 2016; doi:10.1038/leu.2016.260.
Fiskus W, Verstovsek S, Manshouri T, Smith JE, Peth K, Abhyankar S et al. Dual PI3K/AKT/mTOR inhibitor BEZ235 synergistically enhances the activity of JAK2 inhibitor against cultured and primary human myeloproliferative neoplasm cells. Mol Cancer Ther 2013; 12: 577–588.
Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol 2015; 22: 755–763.
Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S et al. Phthalidimide conjugation as a strategy for in vivo target protein degradation. Science 2015; 348: 1376–1381.
Toure M, Crews CM . Small-molecule PROTACS: new approaches to protein degradation. Agnew Chem Int Ed Engl 2016; 55: 1966–1973.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B . Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 7: 621–628.
Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther 2014; 13: 1142–1154.
Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C et al. BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD. Mol Cancer Ther 2014; 13: 2315–2327.
Kornblau SM, Tibes R, Qiu YH, Chen W, Kantarjian HM, Andreeff M et al. Functional proteomic profiling of AML predicts response and survival. Blood 2009; 113: 154–164.
Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP . Mass cytometric functional profiling of acute myeloid leukemia defines cell-cycle and immunophenotypic properties that correlate with known responses to therapy. Cancer Discov 2015; 5: 988–1003.
Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011; 332: 687–696.
Wyspiańska BS, Bannister AJ, Barbieri I, Nangalia J, Godfrey A, Calero-Nieto FJ et al. BET protein inhibition shows efficacy against JAK2V617F-driven neoplasms. Leukemia 2014; 28: 88–97.
Devaraj SG, Fiskus W, Shah B, Qi J, Sun B, Iyer SP et al. HEXIM1 induction is mechanistically involved in mediating anti-AML activity of BET protein bromodomain antagonist. Leukemia 2016; 30: 504–508.
Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 2015; 525: 543–547.
Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 2015; 525: 538–542.
Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 2013; 498: 246–250.
Qui P, Simonds EF, Bendall SC, Gibbs KD, Bruggner RV, Linderman MD et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 2011; 29: 886–891.
Chou TC, Talalay P . Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984; 22: 27–55.
Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010; 141: 69–80.
Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 2014; 21: 1047–1057.
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA et al. Super enhancers in the control of cell identity and disease. Cell 2013; 155: 934–947.
Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS et al. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell 2015; 60: 847–859.
Bhagwat AS, Roe JS, Mok BY, Hohmann AF, Shi J, Vakoc CR . BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep 2016; 15: 519–530.
Levine M, Cattoglio C, Tjian R . Looping back to leap forward: transcription enters a new era. Cell 2014; 157: 13–25.
Lee TI, Young RA . Transcriptional regulation and its misregulation in disease. Cell 2013; 152: 1237–1251.
Blanco-Aparicio C, Carnero A . Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol 2013; 85: 629–643.
Warfel NA, Kraft AS . PIM kinase (and Akt) biology and signaling in tumors. Pharmacol Ther 2015; 151: 41–49.
Dang CV . MYC on the path to cancer. Cell 2012; 149: 22–35.
Li Y, Choi PS, Casey SC, Dill DL, Felsher DW . MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell 2014; 26: 262–272.
Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA 2016; 113: 7124–7129.
Acknowledgements
We thank the Flow Cytometry and Cellular Imaging (FCCI) Core Facility and the Functional Proteomics Reverse-Phase Protein Array (RPPA) Core facility, which are supported by MD Anderson Cancer Center Support Grant 5P30 CA016672-40. The heat maps were developed by the MD Anderson Cancer Center Department of Bioinformatics and Computational Biology, In Silico Solutions, Santeon and SRA International. This work was supported in part by U.S. National Cancer Institute (NCI; MD Anderson TCGA Genome Data Analysis Center) grant numbers CA143883 and CA083639, the Mary K. Chapman Foundation, the Michael & Susan Dell Foundation (honoring Lorraine Dell), and MD Anderson Cancer Center Support Grant P30 CA016672 (the Bioinformatics Shared Resource). Additional support was also provided by CPRIT Metabolomics Core Facility Support Award RP120092 (CC) and a pilot grant from the Alkek Center for Molecular Discovery (CC). CMC acknowledges support from the National Institutes of Health (grant number R35CA197589). This research is supported in part by the MD Anderson Cancer Center Leukemia SPORE (P50 CA100632).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
CMC is the founder and Chief Scientific Advisor of, and possesses an equity ownership stake in, Arvinas, LLC. YQ, KR, KGC, APC and AS are Arvinas employees and possess an equity ownership stake in Arvinas. NP serves on the scientific advisory board of Incyte Pharmaceuticals. He has also served as a consultant and received research funding from Novartis Pharmaceuticals. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Saenz, D., Fiskus, W., Qian, Y. et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia 31, 1951–1961 (2017). https://doi.org/10.1038/leu.2016.393
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.393
This article is cited by
-
Beyond canonical PROTAC: biological targeted protein degradation (bioTPD)
Biomaterials Research (2023)
-
PROTAC’ing oncoproteins: targeted protein degradation for cancer therapy
Molecular Cancer (2023)
-
Epigenetic regulation in hematopoiesis and its implications in the targeted therapy of hematologic malignancies
Signal Transduction and Targeted Therapy (2023)
-
Targeting of epigenetic co-dependencies enhances anti-AML efficacy of Menin inhibitor in AML with MLL1-r or mutant NPM1
Blood Cancer Journal (2023)
-
Bromodomain and extraterminal (BET) proteins: biological functions, diseases, and targeted therapy
Signal Transduction and Targeted Therapy (2023)