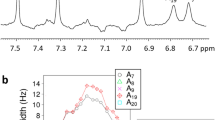
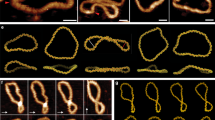
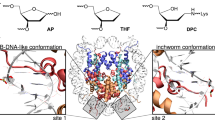
Abstract
Sequence-directed variations in the canonical DNA double helix structure that retain Watson–Crick base-pairing have important roles in DNA recognition, topology and nucleosome positioning. By using nuclear magnetic resonance relaxation dispersion spectroscopy in concert with steered molecular dynamics simulations, we have observed transient sequence-specific excursions away from Watson–Crick base-pairing at CA and TA steps inside canonical duplex DNA towards low-populated and short-lived A•T and G•C Hoogsteen base pairs. The observation of Hoogsteen base pairs in DNA duplexes specifically bound to transcription factors and in damaged DNA sites implies that the DNA double helix intrinsically codes for excited state Hoogsteen base pairs as a means of expanding its structural complexity beyond that which can be achieved based on Watson–Crick base-pairing. The methods presented here provide a new route for characterizing transient low-populated nucleic acid structures, which we predict will be abundant in the genome and constitute a second transient layer of the genetic code.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Watson, J. D. & Crick, F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953)
Record, M. T., Jr et al. Double helical DNA: conformations, physical properties, and interactions with ligands. Annu. Rev. Biochem. 50, 997–1024 (1981)
Koudelka, G. B., Mauro, S. A. & Ciubotaru, M. Indirect readout of DNA sequence by proteins: the roles of DNA sequence-dependent intrinsic and extrinsic forces. Prog. Nucleic Acid Res. Mol. Biol. 81, 143–177 (2006)
Rohs, R. et al. The role of DNA shape in protein–DNA recognition. Nature 461, 1248–1253 (2009)
Segal, E. et al. A genomic code for nucleosome positioning. Nature 442, 772–778 (2006)
Saiz, L. & Vilar, J. M. DNA looping: the consequences and its control. Curr. Opin. Struct. Biol. 16, 344–350 (2006)
Richmond, T. J. & Davey, C. A. The structure of DNA in the nucleosome core. Nature 423, 145–150 (2003)
Wang, A. H. et al. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680–686 (1979)
Patikoglou, G. A. et al. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13, 3217–3230 (1999)
Kitayner, M. et al. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nature Struct. Mol. Biol. 17, 423–429 (2010)
Ughetto, G. et al. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 13, 2305–2323 (1985)
Seaman, F. C. & Hurley, L. Interstrand cross-linking by bizelesin produces a Watson-Crick to Hoogsteen base-pairing transition region in d(CGTAATTACG)2. Biochemistry 32, 12577–12585 (1993)
Yang, H., Zhan, Y., Fenn, D., Chi, L. M. & Lam, S. L. Effect of 1-methyladenine on double-helical DNA structures. FEBS Lett. 582, 1629–1633 (2008)
Shanmugam, G., Kozekov, I. D., Guengerich, F. P., Rizzo, C. J. & Stone, M. P. Structure of the 1,N2-ethenodeoxyguanosine adduct opposite cytosine in duplex DNA: Hoogsteen base pairing at pH 5.2. Chem. Res. Toxicol. 21, 1795–1805 (2008)
Palmer, A. G., III NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 104, 3623–3640 (2004)
Korzhnev, D. M. & Kay, L. E. Probing invisible, low-populated states of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc. Chem. Res. 41, 442–451 (2008)
Boehr, D. D., Nussinov, R. & Wright, P. E. The role of dynamic conformational ensembles in biomolecular recognition. Nature Chem. Biol. 5, 789–796 (2009)
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007)
Korzhnev, D. M., Religa, T. L., Banachewicz, W., Fersht, A. R. & Kay, L. E. A transient and low-populated protein-folding intermediate at atomic resolution. Science 329, 1312–1316 (2010)
Johnson, J. E., Jr & Hoogstraten, C. G. Extensive backbone dynamics in the GCAA RNA tetraloop analyzed using 13C NMR spin relaxation and specific isotope labeling. J. Am. Chem. Soc. 130, 16757–16769 (2008)
Hansen, A. L., Nikolova, E. N., Casiano-Negroni, A. & Al-Hashimi, H. M. Extending the range of microsecond-to-millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R1ρ NMR spectroscopy. J. Am. Chem. Soc. 131, 3818–3819 (2009)
Shajani, Z. & Varani, G. 13C relaxation studies of the DNA target sequence for HhaI methyltransferase reveal unique motional properties. Biochemistry 47, 7617–7625 (2008)
Travers, A. A. The structural basis of DNA flexibility. Philos. Transact. A Math. Phys. Eng. Sci. 362, 1423–1438 (2004)
Pardi, A., Morden, K. M., Patel, D. J. & Tinoco, I., Jr Kinetics for exchange of imino protons in the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix and in two similar helices that contain a G•T base pair, d(C-G-T-G-A-A-T-T-C-G-C-G), and an extra adenine, d(C-G-C-A-G-A-A-T-T-C-G-C-G). Biochemistry 21, 6567–6574 (1982)
Guéron, M., Kochoyan, M. & Leroy, J. L. A single mode of DNA base-pair opening drives imino proton exchange. Nature 328, 89–92 (1987)
Pérez, A., Luque, F. J. & Orozco, M. Dynamics of B-DNA on the microsecond time scale. J. Am. Chem. Soc. 129, 14739–14745 (2007)
Coman, D. & Russu, I. M. A nuclear magnetic resonance investigation of the energetics of basepair opening pathways in DNA. Biophys. J. 89, 3285–3292 (2005)
Song, K. et al. An improved reaction coordinate for nucleic acid base flipping studies. J. Chem. Theory Comput. 5, 3105–3113 (2009)
Greene, K. L., Wang, Y. & Live, D. Influence of the glycosidic torsion angle on 13C and 15N shifts in guanosine nucleotides: investigations of G-tetrad models with alternating syn and anti bases. J. Biomol. NMR 5, 333–338 (1995)
Xu, X. & Au-Yeung, S. Investigation of chemical shift and structure relationships in nucleic acids using NMR and density functional theory methods. J. Phys. Chem. B 104, 5641–5650 (2000)
Izatt, R. M., Christensen, J. J. & Rytting, J. H. Sites and thermodynamic quantities associated with proton and metal ion interaction with ribonucleic acid, deoxyribonucleic acid, and their constituent bases, nucleosides, and nucleotides. Chem. Rev. 71, 439–481 (1971)
Ghosal, G. & Muniyappa, K. Hoogsteen base-pairing revisited: resolving a role in normal biological processes and human diseases. Biochem. Biophys. Res. Commun. 343, 1–7 (2006)
Abrescia, N. G., Thompson, A., Huynh-Dinh, T. & Subirana, J. A. Crystal structure of an antiparallel DNA fragment with Hoogsteen base pairing. Proc. Natl Acad. Sci. USA 99, 2806–2811 (2002)
Nair, D. T., Johnson, R. E., Prakash, L., Prakash, S. & Aggarwal, A. K. Human DNA polymerase ι incorporates dCTP opposite template G via a G.C+ Hoogsteen base pair. Structure 13, 1569–1577 (2005)
Powell, S. W., Jiang, L. & Russu, I. M. Proton exchange and base pair opening in a DNA triple helix. Biochemistry 40, 11065–11072 (2001)
Sau, A. K. et al. Evidence for A+(anti)-G(syn) mismatched base-pairing in d-GGTAAGCGTACC. FEBS Lett. 377, 301–305 (1995)
Lu, L., Yi, C., Jian, X., Zheng, G. & He, C. Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system. Nucleic Acids Res. 38, 4415–4425 (2010)
Gilbert, D. E., van der Marel, G. A., van Boom, J. H. & Feigon, J. Unstable Hoogsteen base pairs adjacent to echinomycin binding sites within a DNA duplex. Proc. Natl Acad. Sci. USA 86, 3006–3010 (1989)
Hoopes, B. C., LeBlanc, J. F. & Hawley, D. K. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J. Mol. Biol. 277, 1015–1031 (1998)
Meyer, T., Carlstedt-Duke, J. & Starr, D. B. A weak TATA box is a prerequisite for glucocorticoid-dependent repression of the osteocalcin gene. J. Biol. Chem. 272, 30709–30714 (1997)
Fischer, S. & Karplus, M. Conjugate peak refinement: an algorithm for finding reaction paths and accurate transition-states in systems with many degrees of freedom. Chem. Phys. Lett. 194, 252–261 (1992)
Segers-Nolten, G. M., Sijtsema, N. M. & Otto, C. Evidence for Hoogsteen GC base pairs in the proton-induced transition from right-handed to left-handed poly(dG-dC)·poly(dG-dC). Biochemistry 36, 13241–13247 (1997)
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995)
Palmer, A. G., III & Massi, F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 106, 1700–1719 (2006)
Miloushev, V. Z. & Palmer, A. G., III R1ρ relaxation for two-site chemical exchange: general approximations and some exact solutions. J. Magn. Reson. 177, 221–227 (2005)
Ferry, J. D., Grandine, L. D. & Fitzgerald, E. R. The relaxation distribution function of polyisobutylene in the transition from rubber-like to glass-like behavior. J. Appl. Phys. 24, 911–916 (1953)
Denisov, V. P., Peters, J., Horlein, H. D. & Halle, B. Using buried water molecules to explore the energy landscape of proteins. Nature Struct. Biol. 3, 505–509 (1996)
Olson, W. K. et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 313, 229–237 (2001)
Abrescia, N. G., Gonzalez, C., Gouyette, C. & Subirana, J. A. X-ray and NMR studies of the DNA oligomer d(ATATAT): Hoogsteen base pairing in duplex DNA. Biochemistry 43, 4092–4100 (2004)
Aishima, J. et al. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 30, 5244–5252 (2002)
MacKerell, A. D., Jr, Banavali, N. & Foloppe, N. Development and current status of the CHARMM force field for nucleic acids. Biopolymers 56, 257–265 (2000)
Chocholoušová, J. & Feig, M. Implicit solvent simulations of DNA and DNA-protein complexes: agreement with explicit solvent vs experiment. J. Phys. Chem. B 110, 17240–17251 (2006)
Feig, M. et al. Performance comparison of generalized born and Poisson methods in the calculation of electrostatic solvation energies for protein structures. J. Comput. Chem. 25, 265–284 (2004)
Lee, M. S., Feig, M., Salsbury, F. R., Jr & Brooks, C. L., III New analytic approximation to the standard molecular volume definition and its application to generalized Born calculations. J. Comput. Chem. 24, 1348–1356 (2003)
Nosé, S. A unified formulation of the constant temperature molecular-dynamics methods. J. Chem. Phys. 81, 511–519 (1984)
Hoover, W. G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 (1985)
Frisch, M. J. et al. Gaussian 03, Revision C.02. (Gaussian, 2004)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Acknowledgements
We thank A. L. Hansen, S. Horowitz and J. Feigon for valuable discussions and suggestions, A. V. Kurochkin for NMR expertise, and C.L. Brooks III for access to a supercomputing cluster. We gratefully acknowledge the Michigan Economic Development Cooperation and the Michigan Technology Tri-Corridor for support in the purchase of a 600 MHz spectrometer. This work was supported by NSF CAREER awards (MCB 0644278 to HMA and CHE-0918817 to I.A.) and an NIH grant (R01GM089846). E.N.N. acknowledges support by a Rackham International and Predoctoral Fellowship awarded by the University of Michigan.
Author information
Authors and Affiliations
Contributions
E.N.N. prepared DNA samples assisted by A.A.W. and performed/analysed all NMR experiments and DFT calculations; E.N.N. and H.M.A. conceived the idea of an HG excited state base pair and approaches to investigate its formation; I.A. and E.K. performed and analysed the MD/CPR simulations; P.J.O. provided expertise and guidance for damaged DNA studies along with critical manuscript revisions; H.M.A. and E.N.N. with help from P.J.O, E.K. and I.A. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-8 with legends, Supplementary Tables 1-6, a Supplementary Discussion, Supplementary References and legends for Supplementary Movies 1-2. (PDF 5534 kb)
Supplementary Movie 1
This movie shows An A•T WC-to-HG base pair transition in duplex B-DNA (see Supplementary Information file for full legend). (MOV 625 kb)
Supplementary Movie 2
This movie shows A G•C WC-to-HG base pair transition in duplex B-DNA (see Supplementary Information file for full legend). (MOV 503 kb)
Rights and permissions
About this article
Cite this article
Nikolova, E., Kim, E., Wise, A. et al. Transient Hoogsteen base pairs in canonical duplex DNA. Nature 470, 498–502 (2011). https://doi.org/10.1038/nature09775
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09775
This article is cited by
-
Dynamic basis for dA•dGTP and dA•d8OGTP misincorporation via Hoogsteen base pairs
Nature Chemical Biology (2023)
-
Experimental detection of conformational transitions between forms of DNA: problems and prospects
Biophysical Reviews (2023)
-
Rational design of hairpin RNA excited states reveals multi-step transitions
Nature Communications (2022)
-
Synthesis and biological evaluation of new aza-acyclic nucleosides and their hydrogen complexes from indole
Research on Chemical Intermediates (2022)
-
Mutate-and-chemical-shift-fingerprint (MCSF) to characterize excited states in RNA using NMR spectroscopy
Nature Protocols (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.