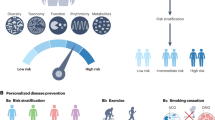
Abstract
Rapid advances in DNA sequencing, metabolomics, proteomics and computational tools are dramatically increasing access to the microbiome and identification of its links with disease. In particular, time-series studies and multiple molecular perspectives are facilitating microbiome-wide association studies, which are analogous to genome-wide association studies. Early findings point to actionable outcomes of microbiome-wide association studies, although their clinical application has yet to be approved. An appreciation of the complexity of interactions among the microbiome and the host's diet, chemistry and health, as well as determining the frequency of observations that are needed to capture and integrate this dynamic interface, is paramount for developing precision diagnostics and therapies that are based on the microbiome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Manichanh, C. et al. Anal gas evacuation and colonic microbiota in patients with flatulence: effect of diet. Gut 63, 401–408 (2014).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007). This study linked the microbiota to IBD and also demonstrated that various forms of the condition have distinct signatures of microbiota.
Lewis, J. D. et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe 18, 489–500 (2015).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Koeth, R. A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature Med. 19, 576–585 (2013).
Kostic, A. D. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 (2012). This study identified high levels of Fusobacterium nucleatum in tissue from human tumours; the bacterium was later confirmed to cause tumours in experiments in animals.
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013). This paper provided the first evidence to directly link the gut microbiota to rheumatoid arthritis in people.
Naseribafrouei, A. et al. Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil. 26, 1155–1162 (2014).
Scheperjans, F. et al. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov. Disord. 30, 350–358 (2015).
Kang, D. W. et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 8, e68322 (2013).
Kostic, A. D., Howitt, M. R. & Garrett, W. S. Exploring host–microbiota interactions in animal models and humans. Genes Dev. 27, 701–718 (2013).
Barr, J. J. et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl Acad. Sci. USA 110, 10771–10776 (2013).
Haiser, H. J. et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013). This study provided a mechanism to underpin the high variation between individuals in efficacy of the cardiac drug digoxin, which was suspected (but not yet proven) to be linked to its metabolism by Eggerthella lenta.
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Knights, D. et al. Bayesian community-wide culture-independent microbial source tracking. Nature Methods 8, 761–763 (2011).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392 (2014). This study of treatment-naive children who had been freshly diagnosed with Crohn's disease enabled the effects of treatment to be separated from those of the condition.
Kuczynski, J. et al. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nature Methods 7, 813–819 (2010).
Friedman, J. & Alm, E. J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8, e1002687 (2012).
Lovell, D., Pawlowsky-Glahn, V., Egozcue, J. J., Marguerat, S. & Bahler, J. Proportionality: a valid alternative to correlation for relative data. PLoS Comput. Biol. 11, e1004075 (2015).
Knights, D., Parfrey, L. W., Zaneveld, J., Lozupone, C. & Knight, R. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe 10, 292–296 (2011).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Fierer, N. et al. Forensic identification using skin bacterial communities. Proc. Natl Acad. Sci. USA 107, 6477–6481 (2010).
Franzosa, E. A. et al. Identifying personal microbiomes using metagenomic codes. Proc. Natl Acad. Sci. USA 112, E2930–E2938 (2015).
Eren, A. M. et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 4, 1111–1119 (2013). This paper demonstrated how careful analysis of exact 16S rRNA sequences that avoids clustering into OTUs can reveal fine-grained information that can be useful for forensic matching.
Noecker, C. et al. Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation. mSystems 13, http://dx.doi.org/10.1128/mSystems.00013-15 (2015).
Koenigsknecht, M. J. et al. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect. Immun. 83, 934–941 (2015).
Hsiao, E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013). This study showed that the phenotype of a mouse model of autism spectrum disorder could be traced, in part, to a single molecule (4-ethylphenylsulfate) and a shift in the microbiota that can be partially restored using a probiotic.
Belda-Ferre, P. et al. The human oral metaproteome reveals potential biomarkers for caries disease. Proteomics 15, 3497–3507 (2015).
Willing, B. P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854 (2010).
Erickson, A. R. et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS ONE 7, e49138 (2012).
Jansson, J. et al. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS ONE 4, e6386 (2009).
Michail, S. et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol. Ecol. 91, 1–9 (2015).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015). This paper compared two discordant studies of microbiomes in type 2 diabetes and showed that the alleged effect of diabetes could be attributed mostly to differences in use of metformin, which has an unexpectedly large effect on the microbiome, between the two populations.
Ridaura, V. K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013). This study demonstrated that phenotypes such as increased adiposity could be transferred from people to mice using personalized culture collections.
Li, H. & Jia, W. Cometabolism of microbes and host: implications for drug metabolism and drug-induced toxicity. Clin. Pharmacol. Ther. 94, 574–581 (2013).
Nicholson, J. K., Lindon, J. C. & Holmes, E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189 (1999).
Wishart, D. S. Emerging applications of metabolomics in drug discovery and precision medicine. Nature Rev. Drug Discov. http://dx.doi.org/10.1038/nrd.2016.32 (2016).
da Silva, R. R., Dorrestein, P. C. & Quinn, R. A. Illuminating the dark matter in metabolomics. Proc. Natl Acad. Sci. USA 112, 12549–12550 (2015).
Gilbert, J. A. & Henry, C. Predicting ecosystem emergent properties at multiple scales. Environ. Microbiol. Rep. 7, 20–22 (2015).
Allen, L. et al. Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. J. Immunol. 174, 3643–3649 (2005).
Puertollano, E., Kolida, S. & Yaqoob, P. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr. Opin. Clin. Nutr. Metab. Care 17, 139–144 (2014).
Cimermancic, P. et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014).
Donia, M. S. et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014). This paper showed that the human microbiome harbours many biosynthetic gene clusters, including those required for the production of antibiotics.
Cummings, J. H. Fermentation in the human large intestine: evidence and implications for health. Lancet 1, 1206–1209 (1983).
Huda-Faujan, N. et al. The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem. J. 4, 53–58 (2010).
Ríos-Covián, D. et al. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7, 185 (2016).
Wikoff, W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. USA 106, 3698–3703 (2009).
Donia, M. S. & Fischbach, M. A. Small molecules from the human microbiota. Science 349, 1254766 (2015).
Nougayrède, J. P. et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006).
Putze, J. et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect. Immun. 77, 4696–4703 (2009).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Langille, M. G. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol. 31, 814–821 (2013).
Allegretti, J. R. et al. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther. 43, 1142–1153 (2016).
Maurice, C. F., Haiser, H. J. & Turnbaugh, P. J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013).
Mani, S., Boelsterli, U. A. & Redinbo, M. R. Understanding and modulating mammalian-microbial communication for improved human health. Annu. Rev. Pharmacol. Toxicol. 54, 559–580 (2014).
Wallace, B. D. et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010). This paper demonstrated that the cancer therapeutic drug irinotecan causes severe diarrhoea because of its reactivation and metabolism by bacterial β-glucuronidases; inhibiting these enzymes with a drug that targets the bacteria, rather than the host, reduces toxicity.
ElRakaiby, M. et al. Pharmacomicrobiomics: the impact of human microbiome variations on systems pharmacology and personalized therapeutics. OMICS 18, 402–414 (2014).
Clayton, T. A., Baker, D., Lindon, J. C., Everett, J. R. & Nicholson, J. K. Pharmacometabonomic identification of a significant host–microbiome metabolic interaction affecting human drug metabolism. Proc. Natl Acad. Sci. USA 106, 14728–14733 (2009). This study provided the first link between the toxicity of a drug (in this case, acetaminophen, a widely used analgesic) and microbial metabolism.
Wilson, I. D. Drugs, bugs, and personalized medicine: pharmacometabonomics enters the ring. Proc. Natl Acad. Sci. USA 106, 14187–14188 (2009).
Quinn, R. A. et al. From sample to multi-omics conclusions in under 48 hours. mSystems http://dx.doi.org/10.1128/mSystems.00038-16 (2016).
Lax, S. et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014).
Gibbons, S. M. et al. Ecological succession and viability of human-associated microbiota on restroom surfaces. Appl. Environ. Microbiol. 81, 765–773 (2015).
Korem, T. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015).
Quick, J. et al. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol. 16, 114 (2015).
Dominguez-Bello, M. G. et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nature Med. 22, 250–253 (2016).
Goyal, M. S., Venkatesh, S., Milbrandt, J., Gordon, J. I. & Raichle, M. E. Feeding the brain and nurturing the mind: linking nutrition and the gut microbiota to brain development. Proc. Natl Acad. Sci. USA 112, 14105–14112 (2015).
Biteen, J. S. et al. Tools for the microbiome: nano and beyond. ACS Nano 10, 6–37 (2016).
Lima-Mendez, G. et al. Determinants of community structure in the global plankton interactome. Science 348, 1262073 (2015).
Sam Ma, Z. et al. Network analysis suggests a potentially 'evil' alliance of opportunistic pathogens inhibited by a cooperative network in human milk bacterial communities. Sci. Rep. 5, 8275 (2015).
Bäckhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 (2015); erratum 17, 852 (2015).
The Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project: dynamic analysis of microbiome–host omics profiles during periods of human health and disease. Cell Host Microbe 16, 276–289 (2014).
Vázquez-Baeza, Y., Pirrung, M., Gonzalez, A. & Knight, R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2, 16 (2013).
Weingarden, A. et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 3, 10 (2015). This paper introduced animation techniques that revealed the transformation of the whole microbiota during faecal microbiota transplantation for C. difficile infection.
Koenig, J. E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108 (suppl. 1), 4578–4585 (2011).
Lozupone, C. A. et al. Meta-analyses of studies of the human microbiota. Genome Res. 23, 1704–1714 (2013).
Flores, G. E. et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 15, 531 (2014).
Shade, A. et al. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5, e01371-14 (2014).
DiGiulio, D. B. et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl Acad. Sci. USA 112, 11060–11065 (2015).
Koren, O. et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480 (2012).
Wang, Z. et al. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur. Heart J. 35, 904–910 (2014).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015). This study showed that individual glycaemic responses could be predicted using the microbiome; it also revealed that although population averages match conventional glycaemic-index values, the responses of individuals are highly idiosyncratic and dependent on the microbiome.
Teng, F. et al. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe 18, 296–306 (2015).
Huang, S. et al. Predictive modeling of gingivitis severity and susceptibility via oral microbiota. ISME J. 8, 1768–1780 (2014).
Zhang, X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nature Med. 21, 895–905 (2015).
Ding, T. & Schloss, P. D. Dynamics and associations of microbial community types across the human body. Nature 509, 357–360 (2014).
Cotillard, A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588 (2013).
Walters, W. A., Xu, Z. & Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233 (2014).
Kang, D., Shi, B., Erfe, M. C., Craft, N. & Li, H. Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci. Transl. Med. 7, 293ra103 (2015).
Sinha, R., Abnet, C. C., White, O., Knight, R. & Huttenhower, C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 16, 276 (2015).
Sinha, R. et al. Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiol. Biomarkers Prev. 25, 407–416 (2016).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
de Goffau, M. C. et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 62, 1238–1244 (2013).
Giongo, A. et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 5, 82–91 (2011).
Kostic, A. D. et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273 (2015).
Scher, J. U. et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 64, 3083–3094 (2012).
Koren, O. et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl Acad. Sci. USA 108 (suppl. 1), 4592–4598 (2011).
Yin, J. et al. Dysbiosis of gut microbiota with reduced trimethylamine-N-oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J. Am. Heart Assoc. 4, e002699 (2015).
Tang, W. H. et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 116, 448–455 (2015).
Xu, R., Wang, Q. & Li, L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics 16 (suppl. 7), S4 (2015).
Tang, W. H. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013).
Zhu, W. et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165, 111–124 (2016).
Hsiao, E. Y., McBride, S. W., Chow, J., Mazmanian, S. K. & Patterson, P. H. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc. Natl Acad. Sci. USA 109, 12776–12781 (2012).
Liu, Z., DeSantis, T. Z., Andersen, G. L. & Knight, R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 36, e120 (2008).
Gilbert, J. A. et al. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand. Genomic Sci. 3, 243–248 (2010).
Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
Quinn, R. A. et al. Microbial, host and xenobiotic diversity in the cystic fibrosis sputum metabolome. ISME J. 10, 1483–98 (2015).
Ridlon, J. M., Kang, D. J., Hylemon, P. B. & Bajaj, J. S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30, 332–338 (2014).
Gill, S. R. et al. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 (2006). This study provided the first metagenomic gene catalogue of the human gut.
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
White, J. R., Nagarajan, N. & Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5, e1000352 (2009).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Mandal, S. et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26, 27663 (2015).
Knights, D., Costello, E. K. & Knight, R. Supervised classification of human microbiota. FEMS Microbiol. Rev. 35, 343–359 (2011).
Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Parks, B. W. et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17, 141–152 (2013).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Acknowledgements
This work and the work in the authors' laboratories that it describes was supported in part by awards from the US National Institutes of Health, the US Department of Energy, the US National Science Foundation, the Alfred P. Sloan Foundation, the Crohn's and Colitis Foundation of America and the US Office of Naval Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com.reprints.
Rights and permissions
About this article
Cite this article
Gilbert, J., Quinn, R., Debelius, J. et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535, 94–103 (2016). https://doi.org/10.1038/nature18850
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18850
This article is cited by
-
Longitudinal analysis shows how microbiome changes might influence responses to immune checkpoint blockade
Nature Medicine (2024)
-
Effects of water ammonia nitrogen on hemolymph and intestinal microbiota of Litopenaeus vannamei
Advanced Biotechnology (2024)
-
On the holobiont ‘predictome’ of immunocompetence in pigs
Genetics Selection Evolution (2023)
-
Chemotherapy-induced weight gain in early-stage breast cancer: a prospective matched cohort study reveals associations with inflammation and gut dysbiosis
BMC Medicine (2023)
-
A microbial causal mediation analytic tool for health disparity and applications in body mass index
Microbiome (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.