Abstract
The majority of human emerging infectious diseases are zoonotic, with viruses that originate in wild mammals of particular concern (for example, HIV, Ebola and SARS)1,2,3. Understanding patterns of viral diversity in wildlife and determinants of successful cross-species transmission, or spillover, are therefore key goals for pandemic surveillance programs4. However, few analytical tools exist to identify which host species are likely to harbour the next human virus, or which viruses can cross species boundaries5,6,7. Here we conduct a comprehensive analysis of mammalian host–virus relationships and show that both the total number of viruses that infect a given species and the proportion likely to be zoonotic are predictable. After controlling for research effort, the proportion of zoonotic viruses per species is predicted by phylogenetic relatedness to humans, host taxonomy and human population within a species range—which may reflect human–wildlife contact. We demonstrate that bats harbour a significantly higher proportion of zoonotic viruses than all other mammalian orders. We also identify the taxa and geographic regions with the largest estimated number of ‘missing viruses’ and ‘missing zoonoses’ and therefore of highest value for future surveillance. We then show that phylogenetic host breadth and other viral traits are significant predictors of zoonotic potential, providing a novel framework to assess if a newly discovered mammalian virus could infect people.
Similar content being viewed by others
Main
Viral zoonoses are a serious threat to public health and global security, and have caused the majority of recent pandemics in people4, yet our understanding of the factors driving viral diversity in mammals, viral host range, and cross-species transmission to humans remains poor. Recent studies have described broad patterns of pathogen host range1,3 and various host or microbial factors that facilitate cross-species transmission5,7,8, or have focused on factors promoting pathogen and parasite sharing within specific mammalian taxonomic groups including primates9,10,11, bats12,13,14, and rodents12,15—but to date there has been no comprehensive, species-level analysis of viral sharing between humans and all mammals. Here we create, and then analyse, a database of 2,805 mammal–virus associations, including 754 mammal species (14% of global mammal diversity) from 15 orders and 586 unique viral species (every recognized virus found in mammals16) from 28 viral families (Methods). We use these data to test hypotheses on the determinants of viral richness and viral sharing with humans. We fit three inter-related models to elucidate specific components of the process of zoonotic spillover (Extended Data Fig. 1). First, we identify factors that influence total viral richness (that is, the number of unique viral species found in a given host, including those which may have the potential to infect humans). Second, we identify and rank the ecological, phylogenetic and life-history traits that make some species more likely hosts of zoonoses than others. Third, recognizing that not all mammalian viruses will have the biological capacity to infect humans, we identify and rank viral traits that increase the likelihood of a virus being zoonotic.
In examining the raw data, we found that observed viral richness within mammals varies at a host order and viral family level, and is highest for Bunya-, Flavi- and Arenaviruses in rodents; Flavi-, Bunya- and Rhabdoviruses in bats; and Herpesviruses in non-human primates (Extended Data Fig. 2). Of 586 mammalian viruses in our dataset, 263 (44.9%) have been detected in humans, 75 of which are exclusively human and 188 (71.5% of human viruses) zoonotic—defined operationally here as viruses detected at least once in humans and at least once in another mammal species (Methods). The proportion of zoonotic viruses is higher for RNA (159 of 382, 41.6%) than DNA (29 of 205, 14.1%) viruses. The observed number of viruses per wild host species was comparable when averaged across orders, but bats, primates, and rodents had a higher proportion of observed zoonotic viruses compared to other groups of mammals (Fig. 1). Species in other orders (for example, Cingulata, Pilosa, Didelphimorphia, Eulipotyphla) also shared a majority of their observed viruses with humans, but data were limited in these less diverse and poorly studied orders. Several species of domesticated ungulates (orders Cetartiodactyla and Perissodactyla) are outliers for their number of observed viruses, but these species have a relatively low proportion of zoonotic viruses (Fig. 1; Supplementary Discussion).
a, b, Box plots of proportion of zoonotic viruses (a) and total viral richness per species (b), aggregated by order. Data points represent wild (light grey, n = 721) and domestic (dark red, n = 32) mammal species; lines represent median, boxes, interquartile range. Animal silhouettes from PhyloPic. Data based on 2,805 host–virus associations. See Methods for image credits and licensing.
Previous analyses show that zoonotic disease emergence events and human pathogen species richness are spatially correlated with mammal and bird diversity2,17. However, these studies weight all species equally. In reality, the risk of zoonotic viral transmission, or spillover, probably varies among host species owing to differences in underlying viral richness, opportunity for contact with humans, propensity to exhibit clinical signs that exacerbate viral shedding18, other ecological, behavioural and life-history differences5,12,15, and phylogenetic proximity to humans10. We hypothesize that the number of viruses a given mammal species shares with humans increases with phylogenetic proximity to humans and with opportunity for human contact. We used generalized additive models (GAMs) to identify and rank host-specific predictors (ecological, life history, taxonomic, and phylogenetic traits, and a control for research effort) of the number of total and zoonotic viruses in mammals (Methods; Supplementary Table 1).
The best-fit model for total viral richness per wild mammal species explained 49.2% of the total deviance, and included a per-species measure of disease-related research effort, phylogenetically corrected body mass, geographic range, mammal sympatry, and taxonomy (order) (Fig. 2a–e). Not surprisingly, research effort had the strongest effect on the total number of viruses per host, explaining 31.9% of the total deviance for this model (Extended Data Table 1). The remaining 17.3% can be explained by biological factors, a value greater than or comparable to studies examining much narrower groups of mammal hosts10,12,15 (Supplementary Discussion). Mammal sympatry was the second most important predictor of total viral richness (Fig. 2d). Our model selection consistently identified mammal sympatry calculated at a ≥20% area overlap over other thresholds explored (Methods), providing insight into the minimum geographic overlap needed to facilitate viral sharing between hosts. Host geographic range was also significantly associated with increasing total viral richness, although the strength of this effect was low (Fig. 2c). Several mammalian orders, Chiroptera (bats), Rodentia (rodents), Primates, Cetartiodactyla (even-toed ungulates), and Perissodactyla (odd-toed ungulates) listed here in order of relative deviance explained, had a significantly greater mean viral richness than predicted by the other variables (Fig. 2e). This finding highlights these taxa as important targets for global viral discovery in wildlife4, and suggests that traits not captured in our analysis (for example, immunological function, social structure, and other life-history variables) may underlie their capacity to harbour a greater number of viral species. Our models to predict total viral richness were comparable when excluding virus–host associations detected by serology, that is, using the ‘stringent data’, and were robust when validated with random cross-validation tests (Extended Data Table 1; Supplementary Table 2). However, we identified several regions that showed significant bias when cross-validated by excluding mammals from zoogeographic areas, suggesting that there are location-specific factors that remain unexplained in our models (Methods; Supplementary Table 3).
Partial effect plots show the relative effect of each variable included in the best-fit GAM, given the effect of the other variables. Shaded circles represent partial residuals; shaded areas, 95% confidence intervals around mean partial effect. a–e, Best model for total viral richness includes: a, number of disease-related citations per host species (research effort, log); b, phylogenetic eigenvector regression (PVR) of body mass (log); c, geographic range area of each species (log km2); d, number of sympatric mammal species overlapping with at least 20% area of target species range; and e, mammalian orders. f–i, Best model for proportion of zoonoses includes: f, research effort (log); g, phylogenetic distance from humans (cytochrome b tree constrained to the topology of the mammal supertree28); h, ratio of urban to rural human population within species range; and i, three mammalian orders. Bats are the only order with a significantly larger proportion of zoonotic viruses than would be predicted by the other variables in the all-data model. Three additional mammalian orders, and whether or not a species is hunted, improved the overall predictive power of the best zoonotic virus model but were non-significant and are not shown (see Extended Data Table 1).
Our best model to predict the number of zoonotic viruses per wild mammal species explained 82% of the deviance, and included phylogenetic distance from humans, the ratio of urban to rural human population across a species range, host order, whether or not a species is hunted, disease-related research effort, and total viral richness (Extended Data Table 1). A large fraction of the deviance explained is driven by the observed total viral richness per host, supporting the biological assumption that the number of viruses that infect humans scales positively with the size of the potential ‘zoonotic pool’19 in each reservoir host. Removing this contribution by including observed total viral richness per host as an offset, the model explains 33% of the total deviance in the proportion of viruses that are zoonotic (Methods), with 30% of total deviance explained by biological factors (Fig. 2f–i). Some mammalian orders had a significant positive (bats) or negative (two ungulate orders) effect on the proportion of zoonotic viruses (Fig. 2i). A number of previous studies have proposed that bats are special among mammals as reservoir hosts of a large number of recently emerging high-profile zoonoses (for example, SARS, Ebola virus, MERS)12,13,20. Our study tests this hypothesis in the context of all known mammalian viruses and hosts. While other mammalian orders have relatively high proportions of observed zoonoses and others have been poorly studied (Fig. 1a), our model results show that bats are host to a significantly higher proportion of zoonoses than all other mammalian orders after controlling for reporting effort and other predictor variables.
We found that the proportion of zoonotic viruses per species increases with host phylogenetic proximity to humans, and that this relationship is significant even when we removed ‘reverse zoonoses’ primarily associated with transmission from humans to primates (Methods). This is the first time this relationship has been demonstrated using data for all mammals and specifically as a determinant of zoonotic spillover, and is supported by previous taxon-specific studies that have examined host relatedness and parasite/pathogen sharing in primates9,10, bats14 and plants21. The proportion of zoonotic viruses shows some upward drift for mammals that are very phylogenetically distant from humans (Fig. 2g) that may represent an artefact of preferentially screening marsupials for human viruses. While primate species largely drive the phylogenetic effect, our best-fit model excluded the effect of the order Primates as a discrete variable (Fig. 2i), suggesting that continuous variation in phylogenetic distance across primate species is more important, and is significant even when all mammals are included. This finding highlights the need to uncover the mechanism by which phylogeny affects spillover risk, for example, evolutionarily related species sharing host cell receptors and viral binding affinities22,23 and specific viral mutations that may expand host range in related mammal species24.
We tested several measures to estimate human–wildlife contact at a global scale for the 721 wild mammals in our dataset, but only the ratio of urban to rural human population (all data model), the change in human population density, and the change in urban to rural population ratio from 1970–2005 across a species range (stringent data model) were included (Extended Data Table 1). The response curve for urban to rural population suggests that increasing urbanization raises the risk of zoonotic spillover (Fig. 2h), as does increasing human population density and the change in urban to rural population ratio over time. A single global metric of human–wildlife ecological contact did not emerge across models. However, the alternate inclusion of these related variables points to the importance of human–animal contact in defining per-species spillover risk globally, and the need for controlled field experiments and human behavioural risk studies to uncover the mechanisms underlying this risk. Overall, the strength of the effect of phylogenetic proximity was stronger than our proxies for animal–human contact in predicting proportion of zoonoses (30–44% stronger explanatory factor), but both remained significant after controlling for research effort (Extended Data Table 1).
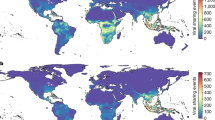
The predominance of zoonoses of wildlife origin in emerging diseases has led to a series of programs to sample wildlife, discover novel viruses, and assess their zoonotic potential4,23,25,26. To inform their scale and scope we calculate the expected number of as-yet undiscovered viruses and zoonoses per host species using our best-fit GAMs and a scenario of increased research effort (Methods, Supplementary Table 4). We then project these ‘missing viruses’ and ‘missing zoonoses’ geographically (Fig. 3, Extended Data Figs 3, 4, 5, 6, 7, 8) to identify regions of the world where targeted, future surveillance to find new viruses and zoonoses will be most effective. In the process of translating our non-spatial, species-level predictions to geographic space, we identified several regions where our model predictions of the number of total and zoonotic viruses were systematically biased (hatched regions in Fig. 3 and Extended Data Figs 3, 4, 5, 6, 7, 8; Methods). Local factors contributing to this bias may include geographic variation in the detection probability of human and/or wildlife viruses, indicating areas where additional research and capacity strengthening for viral detection are most needed. Our model predictions were not systematically biased or clustered across host phylogeny (Extended Data Fig. 9).
Warmer colours highlight areas predicted to be of greatest value for discovering novel zoonotic viruses. a, All wild mammals (n = 584 spp. included in the best-fit model). b, Carnivores (order Carnivora, n = 55). c, Even-toed ungulates (order Cetartiodactyla, n = 70). d, Bats (order Chiroptera, n = 157). e, Primates (order Primates, n = 73). f, Rodents (order Rodentia, n = 183). Hatched regions represent areas where model predictions deviate systematically for the assemblage of species in that grid cell (approximately 18 km × 18 km, see Methods). Animal silhouettes from PhyloPic.
Geographic hotspots of ‘missing zoonoses’ vary by host taxonomic order, with foci for carnivores and even-toed ungulates in eastern and southern Africa, bats in South and Central America and parts of Asia, primates in specific tropical regions in Central America, Africa, and southeast Asia; and rodents in pockets of North and South America and Central Africa. Areas where ‘missing zoonoses’ predictions were systematically biased varied by taxonomic order, but included large parts of Africa for the all-mammal dataset (Fig. 3a, Extended Data Figs 3, 4, 5, 6, 7, 8f). By contrast, the distribution of bias in predicting the ‘missing viruses’ for all mammals was limited to patches of northeastern Asia, Greenland, peninsular Malaysia, and scattered grid cells in western Asia and Patagonia (Extended Data Fig. 3c). We also identify geographic regions with large numbers of mammal species currently lacking any information regarding their viral diversity (Extended Data Figs 3i–8i). In combination, these maps can be used for cost-effective allocation of resources for viral discovery programs, such as the Global Virome Project (D. Carroll et al., submitted).
Finally, a significant challenge to preventing future disease emergence is estimating the zoonotic potential of a newly discovered viral species or strain based on viral traits4,5,6,27. The best model for determining whether or not a known virus (n = 586 mammalian viruses) has been observed as zoonotic explained 27.2% of total deviance and included maximum phylogenetic host breadth (PHB—a virus-specific trait that measures the phylogenetic range of known hosts, excluding humans), research effort, whether or not a virus replicates in the cytoplasm, is vector-borne, or is enveloped, and average genome length (Fig. 4). Using the ‘stringent’ dataset to define whether a virus is zoonotic resulted in a reduced model that excluded enveloped status and genome length (Extended Data Table 1). Our findings confirm a positive relationship between zoonotic potential and ability to replicate in the cytoplasm7, and that viruses with arthropod vectors may be able to infect a wider range of mammalian hosts5. Our phylogenetically explicit measure of host breadth, PHB, can be used at various hierarchical taxonomic levels to quantify and rank viruses from specialist to generalist, and was the strongest predictor of zoonotic potential (12.4% of total deviance explained). This highlights the value of field programs to identify the natural host range of newly discovered pathogens in order to develop early proxies for their zoonotic potential4. Significant variation in PHB across viral families is suggestive of intrinsic differences in the ability of a virus to infect diverse hosts, and this relates to the proportion of observed zoonoses in each family (Fig. 4a).
a, Box plot of maximum phylogenetic host breadth per virus (PHB, see methods) for each of 586 mammalian viruses, aggregated by 28 viral families. Individual points represent viral species, colour-coded by zoonotic status. Box plots coloured and sorted by the proportion of zoonoses in each viral family. b–d, Partial effect plots for the best-fit GAM to predict the zoonotic potential of a virus. b, Maximum PHB. Viruses that infect a phylogenetically broader range of hosts are more likely to be zoonotic. c, Research effort (log, number of PubMed citations per viral species). d, Whether or not a virus replicates in the cytoplasm or is vector-borne. Viral genome length and whether or not a virus is enveloped improved the overall predictive power but were non-significant and are not shown (see Extended Data Table 1).
We acknowledge several important caveats in this study. First, our estimates of missing viruses and missing zoonoses per species are based on the current maximum observed research effort from the literature, and these estimates should be viewed as relative, not absolute. The true size of the undiscovered mammalian virome will probably increase with new genetic tools for unbiased viral discovery and in-depth studies that repeatedly sample wildlife populations over time25. Second, our ecological and biological predictor variables only explain a portion of the total variation in viral richness per host and zoonotic potential based on viral traits, although this is greater than that reported in comparable order-specific studies10,12. Third, while we control for research effort we cannot account for viruses or host associations that have completely evaded human detection to date, nor those identified but not published. Additional resources to support better data sharing and on-the-ground viral surveillance in the species and regions we identify would help validate predictive models to identify zoonotic viral hotspots, and streamline costly efforts to develop measures to prevent their future emergence.
The analyses reported herein have broad potential to assist in expediting viral discovery programs for public health. Our host-specific analyses and estimates of missing zoonoses allow us to identify which species and regions should be preferentially targeted to characterize the global mammalian virome. Our viral trait framework then allows prioritization of newly discovered wildlife viruses for detailed characterization (for example, by sequencing receptor-binding domains, and conducting in vitro and in vivo infection experiments23) to assess their potential to threaten human health.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Database
To construct the mammal–virus association database we initially extracted all viruses listed as occurring in any mammal from the International Committee on Taxonomy of Viruses database (ICTVdb), and further individually went through each virus listed in the ICTV 8th edition master list and searched the literature for mammalian hosts. All viral species names were synonymized to ICTV 8th edition, which was the global authority on viral taxonomy at the start of our data collection in 2010 (ref. 16). From 2010–15 the authors and a team of research assistants and interns at EcoHealth Alliance compiled mammal species associations for each of 586 unique viruses published in the literature between 1940–2015 initially by using the virus name and synonyms as the search keywords in the major online reference databases (Web of Science, PubMed, and Google Scholar) in addition to searching in books, reviews, and literature cited in sources we had already obtained. To narrow the search for hosts for well-researched viruses, we additionally included the terms ‘host(s)’, ‘reservoir’, ‘wildlife’, ‘animals’, ‘surveillance’, and other relevant terms to find publications related to host range. Associations were cross-checked for completeness with the Global Mammal Parasite Database for primate, carnivore and ungulate viruses, version as of Nov 2006 (GMPD, http://www.mammalparasites.org)29 and other published reviews specific to bats and rodents12,30,31. We excluded all records without species-level host information, and those where we could not track down the primary references. Records of mammal–virus associations from experimental infection studies, zoological parks or captive breeding facilities, or cell culture discoveries were excluded. Host species were defined as domestic or wild following the list of domestic animal species from the Food and Agriculture Organization (FAO)32, and we removed the black rat (Rattus rattus) and domestic mouse (Mus musculus) from the domesticated list as these two species make up their own ‘peri-domestic’ category. Host species were categorized as either occurring in human modified habitats or being hunted by humans—both estimates for human contact—according to the IUCN Red List species descriptions33.
To control for the fact that some detection methods are more reliable than others in identifying the pathogen of interest, we recorded the detection method used for each host–virus association and scored these as 0, 1, or 2 according to the reliability of detection method used. Viral isolation and PCR detection with sequence confirmation were scored as a 2 (=stringent data); and serological methods were scored as a 0 or 1, with viral or serum neutralization tests (=1), and enzyme-linked immunoassays (ELISA), antigen detection assays, and other serological assays scored as (=0). ‘Stringent data’ were analysed separately to remove potential uncertainty owing to cross-reactivity with related viruses. We exhaustively searched the literature to identify a stringent detection for each mammal–virus pair, and only included the serological finding for that pair if no molecular or viral isolation studies were available. We partitioned data and conducted separate analyses for the entire data set (0 + 1 + 2 detection quality) and the stringent data (score of 2) to reduce the noise from potential serological cross-reactivity. Full list of host–virus associations, detection methods, and associated references are provided in our data and code repository at http://doi.org/10.5281/zenodo.596810.
Our operational definition of a zoonotic virus includes any virus that was detected in humans and at least one other mammalian host in at least one primary publication, and does not imply directionality. Our complete dataset of mammalian viral associations demonstrates evidence of past or current viral infection which we believe is a reasonable proxy for measuring spillover, and our stringent dataset specifically is more robust to exclude species that may have been exposed to a given virus versus those that show some evidence for replication within the host species. Our bi-directional definition of spillover follows a proposal by the WHO that defines a zoonosis as “any disease or infection that is naturally transmissible from vertebrate animals to humans and vice-versa” (http://www.who.int/zoonoses/en/) and excludes any human pathogens that recently evolved from nonhuman pathogens (for example, HIV in primates), as per Woolhouse and Gowtage-Sequeria (2005) (ref. 1).
In order to address influence of transmission from humans to wildlife in our models, we also ran our GAM model fitting and selection procedure (see below) on a subset of data that excluded any probable ‘reverse zoonotic’ viruses. We first searched our entire dataset and removed any clear instances of transmission from humans to primates, for example, including records from zoological parks and wildlife rehabilitation centres (as previously noted). We then additionally removed several human viruses most commonly transmitted from humans back to non-human primates to create a subset of data without the most common reverse zoonotic viruses (adeno-associated virus-2; human adenovirus D; human herpesvirus 4; human metapneumovirus; human respiratory syncytial virus; measles virus; mumps virus)34,35. We present these additional analyses excluding reverse zoonoses and associated code at http://doi.org/10.5281/zenodo.596810.
Total viral richness was calculated as the number of unique ICTV-recognized viruses found in a given host species, and zoonotic viral richness was defined as the number of unique ICTV-recognized viruses in a given host species that were also detected in humans in our database.
To assess research bias for both host and virus, we searched ISI Web of Knowledge, including Web of Science and Zoological Record, and PubMed for the number of research publications for a given host or pathogen. We recorded two values for the number of research papers for a host. The first was a simple search by scientific binomial in Zoological Abstracts where we recorded the number of papers published between 1940–2013 for each host species. We also recorded the number of disease-related publications for each species using the scientific binomial AND topic keyword: disease* OR virus* OR pathogen* OR parasit*. The * operator was used in our search criteria to capture all words that begin with each term, for example, ‘parasit*’ would return hits for ‘parasite’, ‘parasites’, and ‘parasitic’. These search criteria broadly included papers that examined disease or diseases, virus or viruses, pathogen or pathogens, parasite parasites, or parasitology, for each species. Only one measure of per-host research effort was included at a time in model selection. As these metrics are highly correlated and the number of disease related citations per host outperformed the total number of publications per host in all but one model (all-data zoonoses), we decided to use disease-related publications as our per-species research effort measure for all models to improve interpretability. We also recorded the number of publications for each of 586 virus species using a keyword search by virus name in PubMed and Web of Science. Only one measure of per virus research effort was included at a time in model selection.
We used a phylogenetically corrected measure of body mass (see details below under ‘Phylogenetic signal’) as our main life history predictor variable, because it was the only one for which a nearly complete dataset existed for the species in our dataset. We used the body mass recorded in the PanTHERIA database36 for 709 species. For 3 species, we used the second choice option, body mass recorded in the AnAge database37. For 11 species, we used the third choice option of the extrapolated body mass recorded in PanTHERIA, which is based on body length or forearm length, depending on species. For 36 species, we used the average body mass for members of the genus that had a recorded body mass. We explored other life-history variables related to longevity38, reproductive success, and basal metabolic rate but these were ultimately excluded owing to the high number of missing records.
Phylogenetic signal
We address the issue of non-independence of host species traits owing to shared ancestry39 in our analyses by first quantifying the phylogenetic signal for each variable in our model using Blomberg’s K40. Blomberg’s K measures phylogenetic signal in a given trait by quantifying trait variance relative to an expectation under a Brownian motion null model of evolution using a phylogenetic tree with varying branch lengths. Blomberg’s K-values are scaled from 0 to infinity, with a value of 0 equal to no phylogenetic signal and values greater than 1 equal to strong phylogenetic signal for closely related species that share more similar trait values. While there is no clearly defined K value cut-off in which to apply phylogenetic comparative methods, non-significant value of <1, or more conservatively <0.5, are typical for traits that are phylogenetically independent. The only host variables we examined with significant K values >0.5 were host body mass, and our direct measure of phylogenetic distance to humans. While there are several tools available to control for phylogeny in multivariate analyses, for example, using phylogenetic generalized least square models (for example, PGLS)41, there is currently no modelling approach to control for phylogeny using GAMs. More importantly, a wholesale effort to control for phylogeny across all variables in our analysis was not appropriate here, as we are explicitly testing the relative importance of phylogenetic distance to humans versus other host traits including measures of human–wildlife contact to predict the proportion of zoonotic viruses for a given host species. This left body mass as the only variable in our models, excluding our direct measures of phylogenetic distance, with a significant Blomberg K value that was greater than 1. We controlled for the significant effect of shared evolutionary history using a phylogenetic eigenvector regression (PVR)42,43 on body mass. The PVR approach allowed us to remove phylogenetic signal for any phylogenetically non-independent variables and then include the corrected values back in our GAMs, while retaining predictor variables like phylogenetic distance to humans as unmodified. We calculated PVR for body mass using the R package PVR and our custom-build maximum likelihood host phylogeny using cytochrome b sequences constrained to the order-level topology of the mammalian supertree28,44. Our new variable for body mass that controls for phylogenetic signal (PVRcytb_resid) removed most of the phylogenetic signal, with K = 3.5 unadjusted, and K < 0.5 after PVR correction. Our new metric of body mass scales in the same way, with larger values equal to species with larger body mass. PVR body mass was included in our GAM model selection for the total viral richness and zoonotic virus models.
Host phylogenetic analysis and phylogenetic host breadth
We used two different mammal phylogenetic trees in our analyses and used a model selection framework to determine which best explained our observed association with zoonotic viral richness. First the mammal supertree was pruned in R (package ape, function drop.tips) to include only synonymous species for the 753 species in our database28,45. We synonymized all host species names between the mammal supertree and the host associations in our database using the IUCN Red List33. If the species was listed as ‘cattle’ it was assumed to be Bos taurus, all other records were excluded if there was ambiguity as to the scientific name for the host species. Second, a maximum likelihood cytochrome b tree was generated using the constraint of a multifurcating tree with taxa constrained to their respective orders and the order-level topology matching that of the mammal supertree6, as per this Newick tree file: (MONOTREMATA,((DIDELPHIMORPHIA,(DIPROTODONTIA,PERAMELEMORPHIA)),(PROBOSCIDEA,((PILOSA,CINGULATA),((((RODENTIA,LAGOMORPHA),(PRIMATES,SCANDENTIA)),((((CETARTIODACTYLA,PERISSODACTYLA),CARNIVORA),CHIROPTERA),EULIPOTYPHLA))))))). This generated a higher-resolution species-level mammal tree using cytochrome b data, with more reliable positioning of the higher-level taxonomic relationships than was obtained in exploratory phylogenetic analyses using cytochrome b data alone. GenBank accession numbers and cytochrome b sequence lengths for each species are provided in in our data and code repository. Cytochrome b gene fragments ranged from 143 to 1,140 bp, with >1,000 bp available for 558/665 (84%) of the taxa. Data derived from the cytochrome b tree constrained to the topology of the mammal supertree was selected as the best option in all best-fit GAMs.
Sequences were aligned using MUSCLE with default setting in Geneious R6, and checked visually for errors46. The best maximum likelihood tree with and without the constraint tree were generated using RAxML-HPC2 on XSEDE via the CIPRES Science Gateway server v.3.1 (ref. 47) using a GTR model with parsimony seed, 1.000 bootstrap replicates, and the following, specific parameters (raxmlHPC-HYBRID -s infile -n result -x 12345 -g constraint.tre -N 1000 -c 25 -p 12345 -f a -m GTRCAT).
Matrices of pairwise patristic distances between all species, including Homo sapiens, were calculated from the two phylogenies using the ‘cophenetic’ function in the R package ape45. Phylogenetic trees (Newick format for pruned supertree and cytochrome b tree) and matrices of phylogenetic distance from humans are provided in the data and code repository.
We calculated mean, median, max., min., IQR, and standard deviation (represented as generic function F in equation (1) of phylogenetic host breadth (PHB) from all known mammalian hosts for each virus using the pairwise patristic distances  for each mammal–mammal association for all hosts of a given virus excluding humans, where i indexes each mammal in the database, as does j, and J represents the total mammals in the database. We aggregated these PHB values using mean, median, or maximum values at a viral species, genus and viral family level to generate higher-level taxonomic variables of host breadth per viral group. Our measure is similar to those developed by previous studies to understand parasite host specificity48,49,50, but here we create a generalizable variable to measure viral host breadth that can be aggregated at different viral taxonomic levels.
for each mammal–mammal association for all hosts of a given virus excluding humans, where i indexes each mammal in the database, as does j, and J represents the total mammals in the database. We aggregated these PHB values using mean, median, or maximum values at a viral species, genus and viral family level to generate higher-level taxonomic variables of host breadth per viral group. Our measure is similar to those developed by previous studies to understand parasite host specificity48,49,50, but here we create a generalizable variable to measure viral host breadth that can be aggregated at different viral taxonomic levels.

To make Extended Data Fig. 9, taxon names and terminal branches of cytochrome b tree constrained to supertree were colour-coded using residual from the best-fit zoonotic virus GAM (predicted minus observed zoonotic viral richness) for wildlife species, and plotted using the plot.phylo function in the R package ape45. Symbols (circles) at terminal taxa additionally added to better visualize residual value colours were added using willeerd.nodelabels function (http://dx.doi.org/10.5281/zenodo.10855). All marine mammals, domestic animals, and other taxa with missing data were coded as grey for missing data.
Viral richness heat map (Extended Data Fig. 2) was generated using the R package pheatmap, and the ‘complete’ hierarchical clustering algorithm to sort cells across rows and columns by similar values of viral richness. All box plots, histograms and all other figures generated in R v.3.3.0 (ref. 51). R code for primary figure generation is provided in the code repository.
GAM fitting and selection
We fit a set of generalized additive models (GAMs) that included all of our selected potential variables explaining the number of total viruses or number of zoonoses in hosts, as well as whether viruses were zoonotic (for conceptual framework and summary of each GAM see Extended Data Fig. 1; for full variable list and data sources see Supplementary Table 1). Our use of GAMs, an incorporation of smooth spline predictor functions into the generalized linear model (GLM) framework, allowed us to examine the functional form of our predictor variables (for example, Figs 2 and 4). Categorical and binary variables (for example, host order, IUCN status of hunted or not, and certain viral traits) were fit as random effects of each variable level. We used automated term selection by double penalty smoothing52 to eliminate variables from the models. This method removes variables with little to no predictive power and has been shown to be comparable or superior to comparing alternate models with and without variables. We did use the model comparison method for domestic animals, where the sample size was not sufficient for fitting all variables. In this case dropping variables by double penalty smoothing still allowed pruning the model list to eliminate redundant models. Where there were competing variables measuring the same mechanistic effect, we fit alternate GAMs using only one of each of these variables (as specified in below and in the Extended Data Fig. 1). These included phylogenetic variables, citation counts from alternate databases, and different measures of human population/host overlap. For example, to capture host phylogeny we used phylogenetic distance based on either the mammal supertree20 or a purpose-built cytochrome b constrained by the topology of the mammal supertree, but never both in the same model. For human population variables, we looked at either variables measuring overlap of species range with human-occupied areas, or human population in those areas, as area- and population-based measures were highly co-linear. For citation variables, we looked at either all citations or the number of disease-related citations for each host species, not both, and similarly citations in either PubMed or Web of Knowledge. We used a binomial GAM to analyse the 586 mammalian viruses in our database and identify viral traits that may serve as predictors of zoonotic potential. Co-linearity was not a major issue among variables included in the same model.
We inspected models within 2 AIC units of the model with the lowest AIC, and present the outputs of the best-fit and all other top models (<2 ΔAIC) in our data and code repository. In general, variable effects retained the same functional form and effect size across models within 2 ΔAIC—differences were limited to the adding or dropping of very weak, insignificant effects, or switching between highly correlated competing variables such as citation counts from different databases.
For our model of number of zoonoses per host, we used the total number of observed viruses per host as an offset, effectively fitting a model of proportion of zoonotic viruses per host. We found this variable had a coefficient near to one when it was used as a linear predictor, indicating its appropriateness as an offset.
We repeated the model selection process for all models using the more stringent set of data that used only virus identified in mammal hosts using viral isolation, PCR, or other methods of nucleic acid sequence confirmation, that is, that excluded all associations detected via serology.
All models were fit using the MGCV package for R (version 1.8-12.). We used the model with the lowest AIC to predict the number of expected zoonotic viruses for each host species, using all the data from our database that had complete observations for the best model. Our top models consistently outperform the alternatives by wide margins, as measured by AIC. We used standard methods in the R package MGCV to calculate deviance explained, which is defined as (D_null – D_model)/D_null. In this formula, D_null is the deviance (−2 × likelihood) of an intercept-only, (or, in the case of the zoonoses model, offset-only), model, while D_model is the deviance of our best-fit model.
Analyses were limited to terrestrial mammal species as defined by the IUCN Red List (marine mammals were excluded) and we ran separate analyses for wild and domestic animals. As domestic animals made up a much smaller dataset (n = 32 species) with a unique set of explanatory variables that differed from the wild species analyses, these models were fit separately. Domestic species results are also discussed separately (see Supplementary Discussion) as they are tangential to the primary findings.
Model cross-validation
We used k-fold cross-validation to evaluate goodness of fit for all models. The data was divided into ten folds, selected randomly. For each fold, the model was re-fit based on the other nine folds, and goodness of fit was assessed by conducting a nonparametric permutation test comparing the predicted values versus the real values for the kth fold, where a non-significant result indicates that predictions are unbiased. Poisson models goodness-of-fit may be compared via a parametric χ2 permutation test on deviance values, but this test is inappropriate in the case of models with low mean values, as is our case for some of our GAMs53. The k-fold cross-validation confirmed the robustness of our model predictions for wild mammals, code and outputs from these tests for each best-fit GAM are provided in Supplementary Table 2.
In addition to randomly selected k-fold cross-validation, we evaluated the robustness of our models via a non-random geographic cross-validation, code and summary document provided in our code and data repository. In order to meaningfully organize species in our dataset by geographic areas, we used the 34 zoogeographic regions for terrestrial mammals recently redefined by Holt et al.54. Using QGIS55, a mammal-specific zoogeographical shapefile provided by Holt’s group at the University of Copenhagen (http://macroecology.ku.dk/resources/wallace) was intersected (using QGIS Vector > Geoprocessing Tools > Intersect) with a shapefile of IUCN’s host ranges for all mammals in our database. Areas of these intersections were then calculated using an equal-area projection (Mollweide), and each host was assigned to only the region that contained the greatest proportion of its range. We systematically removed all observations (species) from each given zoogeographical region, re-fit the model using all observations from outside the region, then performed a non-parametric permutation test comparing the predicted values to the observed values for that region. Non-significant results indicate that model predictions are unbiased. Significant results for a given zoogeographic region suggest that there are location-specific biases that remain unexplained. This systematic zoogeographic cross-validation supported the overall robustness of our model predictions for several models, that is, all-data zoonoses, all-data total viral richness, and stringent-data total viral richness models. For these models, even though a majority of zoogeographic regions were unbiased, we still identified several zoogeographic regions that showed significant bias. Our zoogeographic cross-validation was equivocal for the stringent-data zoonoses model, with eight regions that showed evidence of bias and seven regions which showed no evidence of bias (Supplementary Table 3).
The presence of biased regions in our zoogeographic cross-validation suggested the possibility that there is a systematic bias associated with geography not captured by the predictor variables in our models. To further investigate this, we added zoogeographical region as a categorical random effect to each of our best-fit models. For three of our best-fit GAMs (all-data total viruses, stringent-data total viruses, and stringent-data zoonoses) the addition of zoogeographical region as a categorical random effect decreased the model AIC and increased the total deviance explained by 3–5%. The all-data zoonoses model, which was used to create the series of maps in the main manuscript, does not improve with the inclusion of zoogeographical region. However, the improved predictive power of models using region-specific terms is offset by the increase in degrees of freedom (that is, if we included 31 zoogeographic regions as separate terms) and, more importantly, a decreased interpretability of our models—especially when compared to the geographical variables we used, such as host area or species range overlap with human modified habitat. We opted not to include these random effects in our final GAMs in favour of keeping only variables interpretable in the context of our host trait-specific framework. Instead, we indicate areas of geographic bias directly on our spatially mapped outputs. (See ‘Calculating and visualizing missing viruses and missing zoonoses’, below.) Summaries of these models, along with changes in relative deviance explained for the other explanatory variables when zoogeographic region is added as a random effect, are provided in our code and data repository.
Spatial variables
For all the wildlife hosts we used the geographic range information obtained from the IUCN spatial database version 2015.2. Wildlife host species shapefiles needed to replicate analysis are hosted on our Amazon S3 storage (https://s3.amazonaws.com/hp3-shapefiles/Mammals_Terrestrial.zip)33. IUCN depict species’ range distributions as polygons based on the extent of occurrence (EOO), which is defined as the area contained within a minimum convex hull around species’ observations or records. This convex hull or polygon is further improved by including areas known to be suitable or by removing unsuitable or unoccupied areas based on expert knowledge. To accurately calculate the area in km2 of each host species we projected the polygons to an equal area projection (Mollweide).
We calculated various thresholds of mammal sympatry based on percentage of range overlap for each wild species in our database using IUCN shape files for all mammals globally. We define mammal sympatry as the number of mammalian species that overlap with the target species’ geographic range. We calculated mammal sympatry for each wild species in our database at six different thresholds based on the percentage area overlap with the target species geographic range, that is, the number of other wild mammal species with any (>0%), ≥ 20%, ≥ 40%, ≥ 50%, ≥ 80%, or 100% range overlap. The six different thresholds for mammal sympatry were included as competing terms in our model selection for the total viral richness models.
We derived and tested several global measures to estimate the level of human contact with each wild species in our database. To estimate the area of host geographic range covered by crops, pastures, rural and urban areas—as measures of global human contact with a given wildlife species—each species polygon was intersected (overlapped) with spatial data representing those land cover types. Additionally, we calculated the total number of people within each host geographic range using data from HYDE database56, and also separately totalled the number of people in rural and urban populations. We obtained data on the distribution of cropland, pastures, rural and urban areas also from the HYDE database56 for the years 1970, 1980, 1990, 2000 and 2005 with a spatial resolution of 5 × 5 arc minutes, equivalent to 10 km by 10 km at the equator. These datasets were created by combining information from satellite imagery and sub-national crop and pasture statistics56. In our GAMs, we used several transformations of these variables as competing proxies for human–wildlife contact: the log-transformed area of host range that overlapped each type of human-modified land cover, log-transformed human population in the host range, log-transformed human population density in the host range, and the log-ratio of urban and rural human populations in the host range. For each of these, we also included as a variable the change in value from 1970 to 2005. Human–wildlife contact variables that significantly covaried were excluded (set as competing terms) during the model selection process. The ratio of urban to rural human population was used to disentangle variables of human–wildlife contact that significantly covaried. For example, the total area of a species range that overlapped with urban and rural areas was highly correlated with the total geographic area variables we examined (for example, total area, and area in crop, pasture, rural, and urban). The ratio of urban to rural population allowed us to separate these signals and best represent this proxy of per-species human–wildlife contact. All spatial analyses were performed in R (3.3.2)51, using the following R libraries: raster57, rgdal58, and sp58.
Calculating and visualizing missing viruses and missing zoonoses
We used each respective best-fit, all-data GAM from the total viral richness and proportion zoonoses models to calculate the estimated number of viruses that would be observed if the research effort variable for each species was equal to that of the most-studied wild species in our database (Vulpes vulpes with 4,433 total publications and 1,477 disease-related publications). We used the prediction of the total virus richness GAM as the offset for the zoonoses GAM. We then calculated the missing viruses and missing zoonoses by subtracting the observed number of viruses and zoonoses from the predictions based on maximum research for each wild mammalian species.
We used geographic range maps from the IUCN spatial database (2015.2) to visualize the spatial distribution of observed host–virus associations, observed host–zoonoses associations, these associations as predicted under maximum research, and the maximum predicted minus the observed viruses, or the missing viruses and missing zoonoses (for example, Fig. 3; Extended Data Figs 3, 4, 5, 6, 7, 8; Supplementary Table 4). We also generated maps comparing species richness of all species in the IUCN database against those with viral associations in our database. For each species, the distribution range was converted to a grid system with cells 1/6 of a geographic degree (approximately 18 km × 18 km at the equator line). Each grid cell was assigned a value of one to indicate presence. We repeated this process and assigned the observed and predicted-under-maximum-effort number of zoonotic viruses to their correspondent grid cells. Viral and host species richness maps, and both the missing viruses and missing zoonoses maps were calculated by overlying individual grids. Each richness map represents the sum of all values for a given grid cell. We repeated the process for all the host species in our database and created viral and species richness maps for the following orders: Carnivora, Cetartiodactyla, Chiroptera, Primates and Rodentia. These taxa were selected because they represent 681/736 (92.5%) of wild mammal species in our database.
In the process of translating our non-spatial, species-level predictions to geographic space (that is, layered raster maps), we identified several geographic areas where our model predictions of the number of total and zoonotic viruses were systematically biased, that is, P < 0.05 (Supplementary Table 3). In order to visualize the geographic biases of our non-spatial model predictions in our maps (see above regarding zoogeographic cross-validation), we demarcate regions with significant bias with hatching. Hatched regions represent areas where model predictions of total or zoonotic viral richness deviate systematically for the collection of species in that grid cell. For each grid cell we calculated whether the bias exceeded that expected from a random sampling of hosts. This was accomplished by summing the residuals from 100,000 random draws of species in our dataset that was equal to the number of species present in that grid cell, then identifying grid cells where the observed bias was outside the middle 95% of the randomly drawn distribution. We calculated this for all mammals, and separately for each order across all grid cells. Areas with observed bias (outside of 95% of the randomly drawn distribution) are shown with hatched regions on each missing virus and missing zoonoses map.
Animal images used in figures
Animal silhouettes added to Figs 1 and 3 and Extended Data Figs 1 and 2 to visually represent each mammalian order were downloaded from PhyloPic (http://www.phylopic.org). Images used to represent the orders Chiroptera, Cingulata, Diprotodontia, Lagomorpha, Peramelemorphia and Primates were available for use under the Public Domain Dedication license. Images used to represent the orders Carnivora and Rodentia (by R. Groom), Didelphimorphia, Pilosa, and Probscidea (by S. Werning), Eulipotyphyla (by C. Rebler), Certartiodactyla and Perissodactyla (by J. A. Venter, H. H. T. Prins, D. A. Balfour & R. Slotow and vectorized by T. M. Keesey) were provided under a Creative Commons license (https://creativecommons.org/licenses/by/3.0/). We created the silhouette used to represent the order Scandentia.
Data availability
All datasets (host traits, viral traits, full list of host–virus associations and associated references, phylogenetic trees, and phylogenetic distance matrices) needed to fully replicate and evaluate these analyses are provided at http://doi.org/10.5281/zenodo.596810. The top-level README.txt file in the directory details the file structure and metadata provided.
Code availability
All R code and R package dependencies needed to fully replicate and evaluate these analyses are provided at http://doi.org/10.5281/zenodo.596810.
References
Woolhouse, M. E. J. & Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11, 1842–1847 (2005)
Jones, K. E. et al. Global trends in emerging infectious diseases. Nature 451, 990–993 (2008)
Taylor, L. H., Latham, S. M. & Woolhouse, M. E. J. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B 356, 983–989 (2001)
Morse, S. S. et al. Prediction and prevention of the next pandemic zoonosis. Lancet 380, 1956–1965 (2012)
Parrish, C. R. et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72, 457–470 (2008)
Lipsitch, M. et al. Viral factors in influenza pandemic risk assessment. eLife 5, e18491 (2016)
Pulliam, J. R. C. & Dushoff, J. Ability to replicate in the cytoplasm predicts zoonotic transmission of livestock viruses. J. Infect. Dis. 199, 565–568 (2009)
Woolhouse, M. E., Haydon, D. T. & Antia, R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20, 238–244 (2005)
Cooper, N. et al. Phylogenetic host specificity and understanding parasite sharing in primates. Ecol. Lett. 15, 1370–1377 (2012)
Davies, T. J. & Pedersen, A. B. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc. R. Soc. Lond. B 275, 1695–1701 (2008)
Gómez, J. M., Nunn, C. L. & Verdú, M. Centrality in primate-parasite networks reveals the potential for the transmission of emerging infectious diseases to humans. Proc. Natl Acad. Sci. USA 110, 7738–7741 (2013)
Luis, A. D. et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. R. Soc. Lond. B. 280, 20122753 (2013)
Brierley, L., Vonhof, M. J., Olival, K. J., Daszak, P. & Jones, K. E. Quantifying global drivers of zoonotic bat viruses: a process-based perspective. Am. Nat. 187, E53–E64 (2016)
Streicker, D. G. et al. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 329, 676–679 (2010)
Han, B. A., Schmidt, J. P., Bowden, S. E. & Drake, J. M. Rodent reservoirs of future zoonotic diseases. Proc. Natl Acad. Sci. USA 112, 7039–7044 (2015)
Fauquet, C ., Mayo, M. A ., Maniloff, J ., Desselberger, U. & Ball, L. A. Virus taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. (Elsevier Academic Press, 2005)
Dunn, R. R ., Davies, T. J ., Harris, N. C. & Gavin, M. C. Global drivers of human pathogen richness and prevalence. Proc. R. Soc. Lond. B 277, 2587–2595 (2010)
Levinson, J. et al. Targeting surveillance for zoonotic virus discovery. Emerg. Infect. Dis. 19, 743–747 (2013)
Morse, S. S. in Emerging Viruses (ed. Morse, S. S. ) 10–28 (Oxford University Press, 1993)
Zhou, P. et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl Acad. Sci. USA 113, 2696–2701 (2016)
Parker, I. M. et al. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520, 542–544 (2015)
Longdon, B., Brockhurst, M. A., Russell, C. A., Welch, J. J. & Jiggins, F. M. The evolution and genetics of virus host shifts. PLoS Pathog. 10, e1004395 (2014)
Ge, X.-Y. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538 (2013)
Organtini, L. J., Allison, A. B., Lukk, T., Parrish, C. R. & Hafenstein, S. Global displacement of canine parvovirus by a host-adapted variant: A structural comparison between pandemic viruses with distinct host ranges. J. Virol. 89, 1909–1912 (2015)
Anthony, S. J. et al. A strategy to estimate unknown viral diversity in mammals. MBio 4, e00598–13 (2013)
Drexler, J. F. et al. Bats host major mammalian paramyxoviruses. Nat. Commun. 3, 796 (2012)
Geoghegan, J. L., Senior, A. M., Di Giallonardo, F. & Holmes, E. C. Virological factors that increase the transmissibility of emerging human viruses. Proc. Natl Acad. Sci. USA 113, 4170–4175 (2016)
Fritz, S. A., Bininda-Emonds, O. R. P. & Purvis, A. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549 (2009)
Nunn, C. L. & Altizer, S. M. The global mammal parasite database: An online resource for infectious disease records in wild primates. Evol. Anthropol. 14, 1–2 (2005)
Olival, K. J., Epstein, J. H., Wang, L. F., Field, H. E. & Daszak, P. in New Directions in Conservation Medicine: Applied Cases of Ecological Health (eds Aguirre, A. A., Ostfeld, R. S. & Daszak, P. ) Ch. 14, 195–212 (Oxford University Press, 2012)
Calisher, C. H., Childs, J. E., Field, H. E., Holmes, K. V. & Schountz, T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19, 531–545 (2006)
Scherf, B. D. World Watch List for Domestic Animal Diversity. 3rd edn, (Food and Agriculture Organization of the United Nations, 2000)
IUCN. The IUCN Red List of Threatened Species. Version 2014.1, http://www.iucnredlist.org (2014)
Epstein, J. H. & Price, J. T. The significant but understudied impact of pathogen transmission from humans to animals. Mt. Sinai J. Med. 76, 448–455 (2009)
Messenger, A. M., Barnes, A. N. & Gray, G. C. Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom-documented human biological threats to animals. PLoS One 9, e89055 (2014)
Jones, K. E. et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648 (2009)
de Magalhães, J. P. & Costa, J. A database of vertebrate longevity records and their relation to other life-history traits. J. Evol. Biol. 22, 1770–1774 (2009)
Cooper, N., Kamilar, J. M. & Nunn, C. L. Host longevity and parasite species richness in mammals. PLoS One 7, e42190 (2012)
Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (1985)
Blomberg, S. P., Garland, T., Jr & Ives, A. R. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 (2003)
Grafen, A. The phylogenetic regression. Phil. Trans. R. Soc. Lond. B 326, 119–157 (1989)
Diniz-Filho, J. A. F. et al. On the selection of phylogenetic eigenvectors for ecological analyses. Ecography 35, 239–249 (2012)
Diniz-Filho, J. A. F., de Sant’Ana, C. E. R. & Bini, L. M. An eigenvector method for estimating phylogenetic inertia. Evolution 52, 1247–1262 (1998)
Bininda-Emonds, O. R. P. et al. The delayed rise of present-day mammals. Nature 446, 507–512 (2007)
Paradis, E ., Claude, J . & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004)
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004)
Stamatakis, A., Hoover, P. & Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771 (2008)
Cuthill, J. H. & Charleston, M. A. A simple model explains the dynamics of preferential host switching among mammal RNA viruses. Evolution 67, 980–990 (2013)
Poulin, R., Krasnov, B. R. & Mouillot, D. Host specificity in phylogenetic and geographic space. Trends Parasitol. 27, 355–361 (2011)
Poulin, R. & Mouillot, D. Parasite specialization from a phylogenetic perspective: a new index of host specificity. Parasitology 126, 473–480 (2003)
R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ (2014)
Marra, G. & Wood, S. N. Practical variable selection for generalized additive models. Comput. Stat. Data Anal. 55, 2372–2387 (2011)
Pawitan, Y. In All Likelihood: Statistical Modelling and Inference Using Likelihood. (Oxford University Press, 2001)
Holt, B. G. et al. An update of Wallace’s zoogeographic regions of the world. Science 339, 74–78 (2013)
QGIS Geographic Information System. Open Source Geospatial Foundation Project http://www.qgis.org/ (2016)
Goldewijk, K. K., Beusen, A., van Drecht, G. & de Vos, M. The HYDE 3.1 spatially explicit database of human-induced global land-use change over the past 12,000 years. Glob. Ecol. Biogeogr. 20, 73–86 (2011)
raster: Geographic Data Analysis and Modeling version 2.3-40 https://cran.r-project.org/package=raster (2015)
sp: Classes and Methods for Spatial Data version 1.2-1 https://cran.r-project.org/package=sp (2015)
Acknowledgements
This research was supported by the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT program; and NIH NIAID awards R01AI079231 and R01AI110964. The authors thank C. N. Basaraba, J. Baxter, L. Brierley, E. A. Hagan, J. Levinson, E. H. Loh, L. Mendiola, N. Wale and A. R. Willoughby for assistance with data collection, and B. M. Bolker, A. R. Ives, K. E. Jones, C. K. Johnson, A. M. Kilpatrick, J. A. K. Mazet and M. E. J. Woolhouse for comments.
Author information
Authors and Affiliations
Contributions
K.J.O., T.L.B. and P.D. designed the study and supervised the collection of data. N.R., P.R.H. and K.J.O. designed the statistical approach, wrote the code, and generated figures. K.J.O. performed phylogenetic analyses. C.Z.-T. performed spatial analyses. All authors were involved in writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks J. Dushoff and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Conceptual model of zoonotic spillover, viral richness, and summary of models.
a, Conceptual model of zoonotic spillover showing primary risk factors examined, colour-coded according to generalized additive models used. b, Conceptual model of observed, predicted, and actual viral richness in mammals. c, GAMs used in our study to address specific components of a and b, colour-coded by model. Variables listed with ‘or’ under each GAM covaried and were provided as competing terms in model selection, and those in bold were included in the best-fit model using all host–virus associations. Significant variables from each best-fit GAM are noted with an asterisk. Zoonotic viral spillover first depends on the underlying total viral richness in mammal populations and the ecological, taxonomic, and life-history traits that govern this diversity (GAM 1). Second, host- and virus-specific factors may facilitate viral spillover. We examine the relative importance of host phylogenetic distance to humans, ecological opportunity for contact, or other species-specific life-history and taxonomic traits (GAM 2), and identify viral traits associated with a higher likelihood of an observed virus being zoonotic (GAM 3). We estimate the total and zoonotic viral richness per host species using GAMs 1 and 2, and calculate the missing viruses and missing zoonoses under a scenario of increased research effort (b, Methods). Owing to imperfect surveillance in both humans and wildlife and biases in viral detection, there may be uncertainty in the exact proportion of viruses that are zoonotic (b, light grey), and also between the actual, or true, viral richness (dotted lines) and the predicted maximum viral richness per host (dashed line).
Extended Data Figure 2 Heat map of observed total viral richness by mammalian order and viral family.
Dataset includes 754 mammalian species and 586 unique ICTV recognized viral species. Heat map aggregated by rows and columns to group taxa with similar levels of observed viral richness.
Extended Data Figure 3 Global distribution of viral and host species richness for all wild mammals.
a, Observed total viral richness (for n = 576 host spp.); b, predicted total viral richness given maximum research effort; c, missing viruses or predicted minus observed total viral richness; d, observed zoonotic viral richness (n = 584); e, predicted zoonotic viral richness given maximum research effort; f, missing zoonoses or predicted minus observed zoonotic viral richness (same as included in Fig. 3a); g, global mammal species richness (n = 5,290); h, mammal richness for species in our database (n = 753); i, mammal species with no described viruses in the literature. Warmer colours (larger values) in panels c and f highlight areas predicted to be of greatest value for discovering novel viruses or novel viral zoonoses, respectively, in mammals. Red/pink colours in panel i highlight areas with poor viral surveillance in mammal species to date. Hatched regions represent areas where model predictions deviate systematically for the collection of species in that grid cell (see Methods).
Extended Data Figure 4 Global distribution of viral and host species richness for wild carnivores (order Carnivora).
a, Observed total viral richness (for n = 55 host spp.); b, predicted total viral richness given maximum research effort; c, missing viruses or predicted minus observed total viral richness; d, observed zoonotic viral richness (n = 55); e, predicted zoonotic viral richness given maximum research effort; f, missing zoonoses or predicted minus observed zoonotic viral richness (same as included in Fig. 3b); g, global host species richness for Carnivora (n = 276); h, host species richness for Carnivora in our database (n = 79); i, species of the order Carnivora with no described viruses in the literature. Warmer colours (larger values) in c and f highlight areas predicted to be of greatest value for discovering novel viruses or novel viral zoonoses, respectively, in carnivores. Red/pink colours in panel i highlight areas with poor viral surveillance in carnivore species to date. Hatched regions represent areas where model predictions deviate systematically for the collection of species in that grid cell (see Methods).
Extended Data Figure 5 Global distribution of viral and host species richness for wild even-toed ungulates (order Cetartiodactyla).
a, Observed total viral richness (for n = 70 host spp.); b, predicted total viral richness given maximum research effort; c, missing viruses or predicted minus observed total viral richness; d, observed zoonotic viral richness (n = 70); e, predicted zoonotic viral richness given maximum research effort; f, missing zoonoses or predicted minus observed zoonotic viral richness (same as included in Fig. 3c); g, global host species richness for Cetartiodactyla (n = 229); h, host species richness for Cetartiodactyla in our database (n = 105); i, species of the order Cetartiodactyla with no described viruses in the literature. Warmer colours (larger values) in c and f highlight areas predicted to be of greatest value for discovering novel viruses or novel viral zoonoses, respectively, in even-toed ungulates. Red/pink colours in panel i highlight areas with poor viral surveillance in even-toed ungulates species to date. Hatched regions represent areas where model predictions deviate systematically for the collection of species in that grid cell (see Methods).
Extended Data Figure 6 Global distribution of viral and host species richness for bats (order Chiroptera).
a, Observed total viral richness (for n = 156 host spp.); b, predicted total viral richness given maximum research effort; c, missing viruses or predicted minus observed total viral richness; d, observed zoonotic viral richness (n = 157); e, predicted zoonotic viral richness given maximum research effort; f, missing zoonoses or predicted minus observed zoonotic viral richness (same as included in Fig. 3d); g, global host species richness for Chiroptera (n = 1117); h, host species richness for Chiroptera in our database (n = 192); i, species of the order Chiroptera with no described viruses in the literature. Warmer colours (larger values) in c and f highlight areas predicted to be of greatest value for discovering novel viruses or novel viral zoonoses, respectively, in bats. Red/pink colours in panel i highlight areas with poor viral surveillance in bat species to date. Hatched regions represent areas where model predictions deviate systematically for the collection of species in that grid cell (see Methods).
Extended Data Figure 7 Global distribution of viral and host species richness for primates (order Primates).
a, Observed total viral richness (for n = 71 host spp.); b, predicted total viral richness given maximum research effort; c, missing viruses or predicted minus observed total viral richness; d, observed zoonotic viral richness (n = 73); e, predicted zoonotic viral richness given maximum research effort; f, missing zoonoses or predicted minus observed zoonotic viral richness (same as included in Fig. 3e); g, global host species richness for Primates (n = 400); h, host species richness for Primates in our database (n = 98); i, primate species with no described viruses in the literature. Warmer colours (larger values) in c and f highlight areas predicted to be of greatest value for discovering novel viruses or novel viral zoonoses, respectively, in primates. Red/pink colours in panel i highlight areas with poor viral surveillance in primate species to date. Hatched regions represent areas where model predictions deviate systematically for the collection of species in that grid cell (see Methods).
Extended Data Figure 8 Global distribution of viral and host species richness for rodents (order Rodentia).
a, Observed total viral richness (for n = 178 host spp.); b, predicted total viral richness given maximum research effort; c, missing viruses or predicted minus observed total viral richness; d, observed zoonotic viral richness (n = 183); e, predicted zoonotic viral richness given maximum research effort; f, missing zoonoses or predicted minus observed zoonotic viral richness (same as included in Fig. 3f); g, global host species richness for Rodentia (n = 2206); h, host species richness for Rodentia in our database (n = 221); i, rodent species with no described viruses in the literature. Warmer colours (larger values) in c and f highlight areas predicted to be of greatest value for discovering novel viruses or novel viral zoonoses, respectively, in wild rodents. Red/pink colours in panel i highlight areas with poor viral surveillance in rodent species to date. Hatched regions represent areas where model predictions deviate systematically for the collection of species in that grid cell (see Methods).
Extended Data Figure 9 Order-level phylogenies showing residuals from zoonoses model.
a–e, Subtrees from cytochrome b maximum likelihood phylogeny for 558 mammal species (constrained to order-level topology of mammal supertree) for bats (a), carnivores (b), even-toed ungulates (c), rodents (d) and primates (e). Species included have at least one described virus association and available genetic data. Wildlife species names and terminal branches are colour-coded by the residuals (predicted minus observed) from the best-fit GAM to predict the number of zoonotic viruses using all data. Species with residual values between −1 and 1 (black) are accurately predicted within one virus. Warm colours represent species with positive residuals (orange >1 to 3; red >3). Cool colours represent species with negative residuals (green <−1 to −3; blue <−3). Marine mammals, domestic animals, and species with missing data and not included in the best-fit models are shown in grey.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and additional references. (PDF 199 kb)
Supplementary Data
This file contains Supplementary Table 1. This file was added on 23 August 2017. (CSV 17 kb)
Supplementary Data
This file contains Supplementary Table 2. This file was added on 23 August 2017. (CSV 3 kb)
Supplementary Data
This file contains Supplementary Table 3. This file was added on 23 August 2017. (CSV 7 kb)
Supplementary Data
This file contains Supplementary Table 4. This file was added on 23 August 2017. (CSV 82 kb)
Rights and permissions
About this article
Cite this article
Olival, K., Hosseini, P., Zambrana-Torrelio, C. et al. Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017). https://doi.org/10.1038/nature22975
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22975
This article is cited by
-
The impact of anthropogenic climate change on pediatric viral diseases
Pediatric Research (2024)
-
Immunogenetic-pathogen networks shrink in Tome’s spiny rat, a generalist rodent inhabiting disturbed landscapes
Communications Biology (2024)
-
The evolutionary drivers and correlates of viral host jumps
Nature Ecology & Evolution (2024)
-
Zoonotic Parasites in Reptiles, with Particular Emphasis on Potential Zoonoses in Australian Reptiles
Current Clinical Microbiology Reports (2024)
-
Very few scientific publications and newspaper articles focus on catastrophic events and their effects on urban wildlife
Urban Ecosystems (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.