Abstract
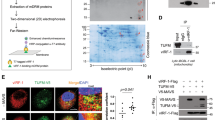
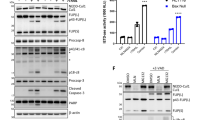
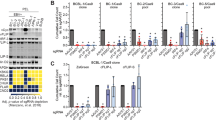
Autophagy is an active homeostatic degradation process for the removal or turnover of cytoplasmic components wherein the LC3 ubiquitin-like protein undergoes an Atg7 E1-like enzyme/Atg3 E2-like enzyme-mediated conjugation process to induce autophagosome biogenesis1,2,3,4. Besides its cytoprotecive role, autophagy acts on cell death when it is abnormally upregulated. Thus, the autophagy pathway requires tight regulation to ensure that this degradative process is well balanced. Two death effector domains (DED1/2) containing cellular FLICE-like inhibitor protein (cFLIP) and viral FLIP (vFLIP) of Kaposi's sarcoma-associated herpesvirus (KSHV), Herpesvirus saimiri (HVS), and Molluscum contagiosum virus (MCV) protect cells from apoptosis mediated by death receptors5,6. Here, we report that cellular and viral FLIPs suppress autophagy by preventing Atg3 from binding and processing LC3. Consequently, FLIP expression effectively represses cell death with autophagy, as induced by rapamycin, an mTor inhibitor and an effective anti-tumour drug against KSHV-induced Kaposi's sarcoma (KS) and primary effusion lymphoma (PEL)7,8. Remarkably, either a DED1 α2-helix ten amino-acid (α2) peptide or a DED2 α4-helix twelve amino-acid (α4) peptide of FLIP is individually sufficient for binding FLIP itself and Atg3, with the peptide interactions effectively suppressing Atg3–FLIP interaction without affecting Atg3-LC3 interaction, resulting in robust cell death with autophagy. Our study thus identifies a checkpoint of the autophagy pathway where cellular and viral FLIPs limit the Atg3-mediated step of LC3 conjugation to regulate autophagosome biogenesis. Furthermore, the FLIP-derived short peptides induce growth suppression and cell death with autophagy, representing biologically active molecules for potential anti-cancer therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008).
Mizushima, N., Levine, B., Cuervo, A. M. & Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Geng, J & Klionsky, D, J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein Modifications: Beyond the Usual Suspects' Review Series. EMBO Rep. 9, 859–864 (2008).
Mizushima, N. Two ubiquitin-like conjugation systems essential for autophagy. Sem. Cell Dev. Biol. 15, 231–236 (2004).
Thome, M. & Tschopp, J. Regulation of lymphocyte proliferation and death by FLIP. Nature Rev. Immunol. 1, 50–58 (2001).
Li, F. Y., Jeffrey, P. D., Yu, J.W & Shi, Y. Crystal structure of a viral FLIP. J. Biol. Chem. 281, 2960–2968 (2006).
Izzedine, H., Brocheriou, I. & Frances, C. Post-transplantation proteinuria and sirolimus. N. Engl. J. Med. 353, 2088–2089 (2005).
Sin, S. H. et al. Rapamycin is efficacious against primary effusion lymphoma (PEL) cell lines in vivo by inhibiting autocrine signaling. Blood 109, 2165–2173 (2007).
Levine, B. & Klionsky, D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 (2004).
Nakatogawa H, Ichimura Y, Ohsumi Y . Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 13, 165–178 (2007).
Ku, B. et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 4, e25 (2008).
Liang, C. et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature Cell. Biol. 8, 688–699 (2006).
Pattingre, S. et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 (2005).
Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T. & Ohsumi, Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 (2004).
Nakamura, H. et al. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77, 4205–4220 (2003).
Guasparri, I., Wu, H. & Cesarman, E. The KSHV oncoprotein vFLIP contains a TRAF-interacting motif and requires TRAF2 and TRAF3 for signalling. EMBO Rep. 7, 114–119 (2006).
Matta, H. & Chaudhary, P. M. Activation of alternative NF-κB pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1β-converting enzyme inhibitory protein (vFLIP). Proc. Natl Acad. Sci. USA 101, 9399–9404 (2004).
Matta, H., Mazzacurati, L., Schamus, S., Yang,.T, Sun, Q & Chaudhary, PM . Kaposi's sarcoma-associated herpesvirus (KSHV) oncoprotein K13 bypasses TRAFs and directly interacts with the IκB kinase complex to selectively activate NF-κB without JNK activation. J. Biol. Chem. 24, 24858–24865 (2007).
Yang, J. K. et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol. Cell 20, 939–949 (2005).
Yamada, Y. et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 282, 8036–8043 (2007).
Bagneris, C. et al. Crystal structure of a vFlip–IKKγ complex: insights into viral activation of the IKK signalosome. Mol. Cell 30, 620–631 (2008).
Ye, F. C. et al. Kaposi's sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-κB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J. Virol. 82, 4235–4249 (2008).
Gump, J. M. & Dowdy, S. F. TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol. Med. 13, 443–448 (2007).
Snyder, E. L., Meade, B. R., Saenz, C.C & Dowdy, S. F. Treatment of terminal peritoneal carcinomatosis by a transducible p53-activating peptide. PLos Biol. 2, 186–193 (2004).
Wadia, J. S., Stan, R.V & Dowdy, S. F. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nature Med. 10, 310–315 (2004).
Hotchkiss, R. S. et al. TAT-BH4 and TAT-Bcl-xL peptides protect against sepsis-induced lymphocyte apoptosis in vivo. J. Immunol. 176, 5471–5477 (2006).
Wu, W., Rochford, R., Toomey, L., Harrington, W. Jr & Feuer, G. Inhibition of HHV-8/KSHV infected primary effusion lymphomas in NOD/SCID mice by azidothymidine and interferon-alpha. Leukemia Res. 29, 545–555 (2005).
Keller, S. A. et al. NF-kappaB is essential for the progression of KSHV- and EBV-infected lymphomas in vivo. Blood 107, 3295–3302 (2006).
Levine, B. Autophagy, immunity, and microbial adaptations. Cell Host Microb. 18, 527–549 (2009).
Mizushima, N., Levine, B., Cuervo, A.M & Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Maiuri, M. C., Zalckvar, E., Kimchi, A & Kroemer, G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature Rev. Mol. Cell Biol. 8, 741–752 (2007).
Scott, R. C., Juhasz, G. & Neufeld, T. P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1–11 (2007).
Rubinztein, D. C., Gestwicki, J. E., Murphy, L.O & Klionsky, D. J. Potential therapeutic applications of autophagy. Nature Rev. Drug Discov. 6, 304–312 (2007).
Acknowledgements
This work was partly supported by US. Public Health Service grants CA82057, CA91819, CA31363, CA115284, AI073099, RR00168, Hastings Foundation, Korean GRL Program K20815000001 (J.J.), AI083841, CA140964, the Lymphoma and Leukemia Society of USA, the Wright Foundation, and the Baxter Foundation (C.L.). We thank Don Ganem, Noboru Mizushima, Preet Chaudhary and Jeff Cohen for reagents, Ernesto Barron for EM analysis, and Steven Lee for manuscript preparation. Finally, we thank all of J.J.'s lab members for their discussions.
Author information
Authors and Affiliations
Contributions
J.S.L. performed all aspects of this study; Q.L. performed the initial study; J.Y.L, S.H.L, H.J, and H.R.L assisted with the experimental design and in collecting the data; F.C.Z and S.J.G. provided KSHV ΔvFLIP; C.L. assisted with the experimental design and interpretation; J.S.L. and J.J. organized this study and wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 4248 kb)
Rights and permissions
About this article
Cite this article
Lee, JS., Li, Q., Lee, JY. et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 11, 1355–1362 (2009). https://doi.org/10.1038/ncb1980
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1980
This article is cited by
-
Delineating the twin role of autophagy in lung cancer
Biologia Futura (2023)
-
c-FLIP regulates autophagy by interacting with Beclin-1 and influencing its stability
Cell Death & Disease (2021)
-
Modulation of virus-induced NF-κB signaling by NEMO coiled coil mimics
Nature Communications (2020)