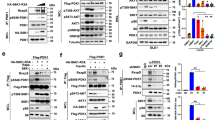
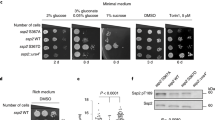
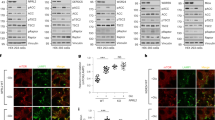
Abstract
The mechanistic target of rapamycin (mTOR) functions as a critical regulator of cellular growth and metabolism by forming multi-component, yet functionally distinct complexes mTORC1 and mTORC2. Although mTORC2 has been implicated in mTORC1 activation, little is known about how mTORC2 is regulated. Here we report that phosphorylation of Sin1 at Thr 86 and Thr 398 suppresses mTORC2 kinase activity by dissociating Sin1 from mTORC2. Importantly, Sin1 phosphorylation, triggered by S6K or Akt, in a cellular context-dependent manner, inhibits not only insulin- or IGF-1-mediated, but also PDGF- or EGF-induced Akt phosphorylation by mTORC2, demonstrating a negative regulation of mTORC2 independent of IRS-1 and Grb10. Finally, a cancer-patient-derived Sin1-R81T mutation impairs Sin1 phosphorylation, leading to hyper-activation of mTORC2 by bypassing this negative regulation. Together, our results reveal a Sin1-phosphorylation-dependent mTORC2 regulation, providing a potential molecular mechanism by which mutations in the mTORC1–S6K–Sin1 signalling axis might cause aberrant hyper-activation of the mTORC2–Akt pathway, which facilitates tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011).
Wullschleger, S., Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Guertin, D. A. & Sabatini, D. M. Defining the role of mTOR in cancer. Cancer Cell 12, 9–22 (2007).
Sabatini, D. M. mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer 6, 729–734 (2006).
Dazert, E. & Hall, M. N. mTOR signaling in disease. Curr. Opin. Cell Biol. 23, 744–755 (2011).
Alayev, A. & Holz, M. K. mTOR signaling for biological control and cancer. J. Cell Physiol. 228, 1658–1664 (2013).
Jewell, J. L., Russell, R. C. & Guan, K. L. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139 (2013).
Guertin, D. A. & Sabatini, D. M. An expanding role for mTOR in cancer. Trends Mol. Med. 11, 353–361 (2005).
Inoki, K., Corradetti, M. N. & Guan, K. L. Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 37, 19–24 (2005).
Kim, D. H. et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002).
Sarbassov, D. D. et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 (2004).
Jacinto, E. et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125–137 (2006).
Frias, M. A. et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 16, 1865–1870 (2006).
Yang, Q., Inoki, K., Ikenoue, T. & Guan, K. L. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832 (2006).
Hung, C. M., Garcia-Haro, L., Sparks, C. A. & Guertin, D. A. mTOR-dependent cell survival mechanisms. Cold Spring Harb. Perspect. Biol. 4, a008771 (2012).
Jacinto, E. et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 (2004).
Ma, X. M. & Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 (2009).
Chan, E. Y. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci. Signal 2, pe51 (2009).
Pelletier, C. L. et al. TSC1 sets the rate of ribosome export and protein synthesis through nucleophosmin translation. Cancer Res. 67, 1609–1617 (2007).
Dibble, C. C. & Manning, B. D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 15, 555–564 (2013).
Efeyan, A., Zoncu, R. & Sabatini, D. M. Amino acids and mTORC1: from lysosomes to disease. Trends Mol. Med. 18, 524–533 (2012).
Inoki, K., Li, Y., Zhu, T., Wu, J. & Guan, K. L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 (2002).
Manning, B. D., Tee, A. R., Logsdon, M. N., Blenis, J. & Cantley, L. C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10, 151–162 (2002).
Sancak, Y. et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25, 903–915 (2007).
Vander Haar, E., Lee, S. I., Bandhakavi, S., Griffin, T. J. & Kim, D. H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9, 316–323 (2007).
Harrington, L. S. et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166, 213–223 (2004).
Shah, O. J., Wang, Z. & Hunter, T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14, 1650–1656 (2004).
Hsu, P. P. et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 (2011).
Yu, Y. et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 (2011).
Pearce, L. R. et al. Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1). Biochem. J. 431, 245–255 (2010).
Gao, D. et al. Rictor forms a complex with Cullin-1 to promote SGK1 ubiquitination and destruction. Mol. Cell 39, 797–808 (2010).
Kim, D. H. et al. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11, 895–904 (2003).
Guertin, D. A. et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 11, 859–871 (2006).
Dibble, C. C., Asara, J. M. & Manning, B. D. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol. Cell Biol. 29, 5657–5670 (2009).
Treins, C., Warne, P. H., Magnuson, M. A., Pende, M. & Downward, J. Rictor is a novel target of p70 S6 kinase-1. Oncogene 29, 1003–1016 (2010).
Romanelli, A., Dreisbach, V. C. & Blenis, J. Characterization of phosphatidylinositol 3-kinase-dependent phosphorylation of the hydrophobic motif site Thr(389) in p70 S6 kinase 1. J. Biol. Chem. 277, 40281–40289 (2002).
Obata, T. et al. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem. 275, 36108–36115 (2000).
Humphrey, S. J. et al. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 (2013).
Gupta, R. K. et al. Transcriptional control of preadipocyte determination by Zfp423. Nature 464, 619–623 (2010).
Huttlin, E. L. et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Zhang, H. et al. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J. Clin. Invest. 117, 730–738 (2007).
Garcia-Martinez, J. M. & Alessi, D. R. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416, 375–385 (2008).
Yan, L., Mieulet, V. & Lamb, R. F. mTORC2 is the hydrophobic motif kinase for SGK1. Biochem. J. 416, e19–e21 (2008).
Purvis, J. E. & Lahav, G. Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 (2013).
Ikeda, F. & Lahav, G. Signal transduction and signaling networks. Mol. Biol. Cell 24, 676 (2013).
Purvis, J. E. & Lahav, G. Decoding the insulin signal. Mol. Cell 46, 715–716 (2012).
Purvis, J. E. et al. p53 dynamics control cell fate. Science 336, 1440–1444 (2012).
Meier, R., Alessi, D. R., Cron, P., Andjelkovic, M. & Hemmings, B. A. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bβ. J. Biol. Chem. 272, 30491–30497 (1997).
Baer, K. et al. Activation of a GST-tagged AKT2/PKBβ. Biochim. Biophys. Acta 1725, 340–347 (2005).
Zhang, X., Tang, N., Hadden, T. J. & Rishi, A. K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta 1813, 1978–1986 (2011).
Paik, J. H. et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128, 309–323 (2007).
Gan, B. et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18, 472–484 (2010).
Vogt, P. K., Jiang, H. & Aoki, M. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle 4, 908–913 (2005).
Brunet, A. et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 (1999).
Network, C. G. A. R. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Forbes, S. A. et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 38, D652–D657 (2010).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Shankaran, H. et al. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol. Syst. Biol. 5, 332 (2009).
Chen, J. Y., Lin, J. R., Cimprich, K. A. & Meyer, T. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Mol. Cell 45, 196–209 (2012).
Gao, D., Inuzuka, H., Tseng, A., Chin, R. Y., Toker, A. & Wei, W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat. Cell Biol. 11, 397–408 (2009).
Chin, Y. R. & Toker, A. The actin-bundling protein palladin is an Akt1-specific substrate that regulates breast cancer cell migration. Mol. Cell 38, 333–344 (2010).
Lin, H. K. et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat. Cell Biol. 11, 420–432 (2009).
Wei, W. et al. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 428, 194–198 (2004).
Boehm, J. S., Hession, M. T., Bulmer, S. E. & Hahn, W. C. Transformation of human and murine fibroblasts without viral oncoproteins. Mol. Cell Biol. 25, 6464–6474 (2005).
Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005).
Campisi, J. & d’Adda di Fagagna, F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007).
Min, S. H. et al. Negative regulation of the stability and tumor suppressor function of fbw7 by the pin1 prolyl isomerase. Mol. Cell 46, 771–783 (2012).
Acknowledgements
We thank A. Toker, J. Guo, K. Xu, A. W. Lau and A. Tron for critical reading of the manuscript, S. J. Elledge (Harvard Medial School, USA), S. Ishii (University of Tsukuba, Japan), W. Hahn (Dana-Farber Cancer Institute, Harvard Medical School, USA), D. Sarbassov (UT M. D. Anderson Cancer Center, USA) K. Guan (UC San Diego, USA) and J. Dempsey (Harvard Medical School, USA) for providing valuable reagents, B. Spiegelman (Dana-Farber Cancer Institute, Harvard Medical School, USA) for providing 3T3-L1 adipocyte cells, L. Cantley and A. Toker for helpful suggestions, and members of the W.W., J.B. and B.S. laboratories for useful discussions. W.W. is an ACS research scholar and a LLS research scholar. Y. Yu is a CPRIT Scholar (CPRIT R1103) in Cancer Research and a Virginia Murchison Linthicum Scholar in Medical Research. P.L. is a NRSA T32 trainee and supported by 5T32HL007893. This work was supported in part by NIH grants (W.W., GM089763, GM094777 and CA177910; and B.S., AI063348 and PR093728).
Author information
Authors and Affiliations
Contributions
P.L., W.G. and H.I. performed most of the experiments with assistance from D.G., L.W., A.S.L., S.S. and O.A. W.W. and B.S. designed the experiments and supervised the study. P.L. and W.W. wrote the manuscript. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Sin1 plays a critical role in negative regulation of Akt-Ser473 phosphorylation in response to PDGF or EGF stimulation independent of Grb10 or IRS-1.
a. Inhibiting mTORC1/S6K by various inhibitors led to elevated Akt-Ser473 phosphorylation. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from HeLa cells that were serum-starved for 24 hours and then collected after insulin stimulation for 30 minutes. Where indicated, the kinase inhibitors (AktVIII: 10 μM, PP242: 1 μM, Rapamycin: 20 nM, S6K1-I: 10 μM) were added together with insulin (100 nM). DMSO was used as a negative control. b. Depletion of endogenous S6K1 or endogenous Raptor led to elevated Akt-Ser473 phosphorylation. IB analysis of WCL derived from HeLa cells infected with the indicated lentiviral shRNA constructs. 24 hours post-infection, cells were selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells. c-d. Depletion of endogenous TSC2, which activates S6K, led to a reduction in Akt-Ser473 phosphorylation. IB analysis of WCL derived from WT or TSC2−/− MEFs (c) or HeLa cells depleted of endogenous TSC2 via lentiviral infections (d). e. Depletion of endogenous Grb10 did not significantly affect Akt-Ser473 phosphorylation in response to PDGF or EGF. TSC2−/− MEFs were infected with the shGrb10 (with shGFP as a negative control) lentiviral construct and selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells. Afterwards, the generated various TSC2−/− MEFs were serum-starved for 24 hours and then collected after stimulation with increasing dose of the indicated stimuli for 30 minutes for IB analysis. f–g. Depletion of endogenous IRS-1 or IRS-1/Grb10 double knockdown did not significantly affect Akt-Ser473 phosphorylation in response to PDGF or EGF. TSC2−/− MEFs were infected with the shIRS-1 (with shGFP as a negative control) lentiviral construct and selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells (f). The IRS-1-depleted TSC2−/− MEFs were subsequently infected with the shGrb10 (with shGFP as a negative control) lentiviral construct to generate IRS-1/Grb10 double-depletion cell lines (g). Afterwards, the generated various TSC2−/− MEFs were serum-starved for 24 hours, stimulated with increasing doses of the indicated stimuli (insulin: 100 nM; IGF-1: 100 ng/ml and PDGF: 100 ng/ml for 30 minutes; or EGF: 100 ng/ml for 10 minutes) then lysed for IB analysis. h-i. IB analysis to demonstrate the IRS-1 (h) or Grb10 (i) depletion efficiency. TSC2−/− MEFs were infected with either shIRS-1 or shGrb10 (with shGFP as a negative control) lentiviral constructs and selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells. Afterwards, the whole cell lysates were collected for IB analysis. j-m. Akt-Ser473 phosphorylation could still be augmented upon S6K1-I (j) or rapamycin (k) inhibitor treatment in cells depleted of both endogenous IRS-1 and Grb10. TSC2−/− MEFs were infected with shIRS-1 or shGrb10 (with shGFP as a negative control) lentiviral shRNA constructs and selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells. The IRS-1-depleted TSC2−/− MEFs were subsequently infected with the shGrb10 (with shGFP as a negative control) lentiviral shRNA construct to generate the IRS-1/Grb10 double-depletion cell lines (l-m). Afterwards, the generated various TSC2−/− MEFs were treated with 10 μM S6K inhibitor (j, l) or 20 nM rapamycin (k, m) for 12 hours before collecting the whole cell lysates for IB analysis. n. WT or Sin1−/− MEFs were serum-starved for 24 hours and then collected after stimulation with the indicated stimuli for 30 minutes for IB analysis. o. Depletion of endogenous Sin1 in TSC2−/− MEFs led to an abolishment of Akt-Ser473 phosphorylation. IB of WCL derived from shGFP- or shSin1-TSC2−/− MFEs that were serum starved for 12 hours and then treated with the indicated stimuli for the indicated time periods before harvesting for IB analysis. Specifically, the doses of stimuli used are listed below: insulin (100 nM), IGF-1 (100 ng/ml), PDGF (100 ng/ml) and EGF (100 ng/ml). Other than the indicated time points for EGF, cells were treated for 30 min with insulin, IGF-1 or PDGF before harvesting.
Supplementary Figure 2 S6K1 is the major physiological kinase responsible for Sin1 phosphorylation at both T86 and T398 sites in a tissue-specific or context-dependent manner.
a. AGC kinases in Fig. 1b were active, as illustrated by their comparable capability to phosphorylate Rictor in cells. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates derived from 293T cells transfected with Myc-Rictor and indicated HA-tagged constitutive active AGC family kinases. b. A dominant negative form of S6K (K/R) mutant could block Sin1 phosphorylation in 293T cells. IB analysis of WCL and immunoprecipitates derived from 293T cells transfected with Flag–Sin1 and HA-S6K1-WT or a constitutive active form of S6K1 in the presence or absence of 20 nM rapamycin. c. Depletion of endogenous TSC2, which led to elevated S6K kinase activity, resulted in increased Sin1 phosphorylation in cells. IB of WCL and Flag immunoprecipitates derived from shGFP- or shTSC2-HeLa cells that were transfected with the Flag–Sin1 construct (pcDNA3 plasmid was used as a negative control). 30 hours post-transfection, cells were serum starved for 24 hours and then treated with insulin for 30 minutes before harvesting for IB analysis. d. Schematic representation of the Sin1 peptide where the T86 site was identified to be phosphorylated in vivo by the mass spectrometry analysis. Please refer to the methods section ‘Mass Spectrometry Analysis’ for experimental details. e. Schematic representation of the Sin1 peptide where the T398 site was identified to be phosphorylated in vitro by S6K1 using the mass spectrometry analysis. Please refer to the methods section “Mass Spectrometry Analysis” for experimental details. f. Characterization of the generated Sin1-pT86 antibody. IB analysis of WCL and Flag immunoprecipitates (IP) derived from 293T cells transfected with HA-S6K1 and the indicated Flag–Sin1 constructs, by blotting with the generated phospho-Sin1-T86 antibody (Sin1-pT86) used in the rest of the studies in this paper. g. Characterization of the generated Sin1-pT398 antibody. IB analysis of Flag-IP derived from 293T cells transfected with HA-S6K1 and the indicated Flag–Sin1 constructs, by blotting with the generated phospho-Sin1-T398 antibody (Sin1-pT398) used in the rest of the studies in this paper. h. Sin1-pT398 signals could be induced by insulin in vivo. IB analysis of Flag-IP derived from HeLa cells transfected with the Flag–Sin1 construct by the generated Sin1-pT398 antibody. Where indicated, 100 nM insulin was added for 30 min prior to cell collection after overnight serum starvation. i. Depletion of endogenous Raptor or endogenous S6K1 resulted in an decrease in Sin1 phosphorylation in vivo. IB analysis of WCL and Flag-IP derived from Flag–Sin1-transfected HeLa cells that were previously infected with the indicated lentiviral shRNA to deplete endogenous Raptor or S6K1. 24 hours post transfection, cells were serum starved for another 24 hours followed by EGF stimulation (100 ng/ml) for 10 minutes before harvesting for IB analysis. j. The S6K1 inhibitor, S6K1-I, could partially block Sin1 phosphorylation induced by insulin, IGF-1, PDGF or EGF. IB analysis of WCL and Flag-IP derived from HeLa cells transfected with Flag–Sin1. 24 hours post transfection, cells were serum starved for another 12 hours followed by treatment with different stimuli (insulin: 100nM, IGF-1: 100 ng/ml, PDGF: 100 ng/ml for 30 minutes or EGF: 100 ng/ml for 10 min) in the presence or absence of 10 μM S6K1 inhibitor (S6K1-I) before harvesting for IB analysis. k. S6K phosphorylates Sin1 on T86 in vitro. GST-Sin1 was incubated with recombinant active S6K protein as described in the in vitro kinase assay section. About 20 ng of the GST-Sin1 was resolved on SDS-PAGE and blotted with the Sin1-pT86 antibody. l. Rapamycin could block WT-S6K, but not the rapamycin resistant form of S6K (CA), in phosphorylating Sin1 in vivo. IB analysis of WCL and Flag-IP derived from 293T cells transfected with Flag–Sin1 and HA-S6K1-WT (WT) or S6K1 (CA) in the presence or absence of 20 nM rapamycin. m. At endogenous levels, Sin1 phosphorylation was attenuated upon mTORC1 or S6K inhibition. IB analysis of WCL derived from foreskin fibroblasts that were serum-starved for 24 hours and then collected after stimulation with 10% FBS for 30 minutes. Where indicated, the indicated kinase inhibitors (AktVIII: 10 μM, PP242: 1 μM, Rapamycin: 20 nM, S6K1-I: 10 μM) were added together with insulin (100 nM). DMSO was used as a negative control. n. Sin1 phosphorylation was markedly decreased when endogenous Akt1 and S6K1 were depleted simultaneously. IB analysis of WCL and Flag-IP derived from Flag–Sin1-transfected HeLa cells depleted of endogenous Akt1, Akt2 or together with S6K1. o. Rapamycin could partially restore Akt-pSer473 in TSC2−/− MEFs in part via reducing Sin1 phosphorylation in vivo. IB analysis of WCL derived from WT or TSC2−/− MEFs. Where indicated, cells were treated with 10 μM S6K inhibitor (S6K1-I) or 20 nM rapamycin before collecting WCL for IB analysis. p. Rapamycin could partially block in vivo Sin1 phosphorylation induced by insulin in primary foreskin human fibroblasts. IB analysis of WCL derived from primary foreskin fibroblasts that were serum starved for 24 hours followed by insulin stimulation (100 nM) for 30 minutes with or without rapamycin (20 nM) before harvesting for IB analysis. q. In 3T3-L1 adipocytes, Akt is the major kinase for Sin1-T86 phosphorylation, while both Akt and S6K are mediating Sin1-T398 phosphorylation. IB analysis of WCL derived from 3T3-L1 cells that were serum starved for 24 hours before addition of 100 nM insulin for 30 min together with the indicated inhibitors (AktVIII: 10 μM, PP242: 1 μM, rapamycin: 20 nM, S6K1-I: 10 μM). r. In HeLa cells, both S6K1 and Akt are involved in phosphorylating Sin1-T86, whereas S6K1 is the major kinase for Sin1-T398 phosphorylation. IB analysis of WCL derived from HeLa cells that were serum starved for 24 hours before addition of 100 nM insulin for 30 min together with the indicated inhibitors (AktVIII: 10 μM, PP242: 1 μM, rapamycin: 20 nM, S6K1-I: 10 μM).
Supplementary Figure 3 Sin1 phosphorylation led to its dissociation from the mTORC2 complex.
a-c. Under ectopic overexpression conditions, Sin1 phospho-mimetic mutant (Sin1-EE) exhibited a reduced interaction with Rictor (a-b) or mTOR (c), indicating that Sin1 phosphorylation led to the dissociation of Sin1 from Rictor or mTOR. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with the indicated Flag–Sin1 constructs and Myc-Rictor (a), HA-Rictor (b) or Myc-mTOR (c). d. Sin1 phosphorylation on both T86 and T398 are necessary to dissociate Sin1 from other mTORC2 components. IB analysis of WCL and IP derived from 293T cells transfected with the indicated Flag–Sin1 constructs (EV: empty vector control; WT: Sin1-WT; EE: Sin1-T86E/T398E). e.Rictor could not form complex with Sin1 phosphorylated on both T86 and T398. Indicated Flag–Sin1 constructs were transfected together with HA-Rictor into Sin1−/− MEFs and 48 hours post transfection, HA-Rictor precipitation was performed and subjected to immunobloting with the indicated antibodies. f. Gel filtration experiments to illustrate that depletion of endogenous TSC2 resulted in a disruption of Sin1 association with mTORC2 in vivo that is associated with elevated Sin1 phosphorylation. IB analysis of the indicated fractionations derived from the gel filtration experiment with Flag–Sin1-transfected HeLa-shGFP or HeLa-shTSC2 WCL harvested in EBC buffer. g. Quantification of total Sin1 abundance illustrated in (f). h-i. Deletion of endogenous TSC2, which led to increased S6K kinase activity, resulted in a reduction of Rictor association with Sin1, which could be partially converted by inhibiting S6K activity by S6K1-I. IB analysis of WCL and anti-Sin1 IP derived from WT or TSC2−/− MEFs. Where indicated, S6K1-I (10 μM) was added 12 hours prior to cell collection.
Supplementary Figure 4 Impairment of Sin1 phosphorylation led to stabilized mTORC2 integrity and subsequently sustained Akt activation under various stimulation conditions.
a–b. Quantification curves for the relative pT86-Sin1 intensity and Rictor abundance as a function to the treatment time as presented in Fig. 3a,b. c,d. Rapamycin treatment attenuated in vivo Sin1 phosphorylation and a subsequent dissociation of Sin1 from Rictor/mTOR under insulin stimulation condition (c) or EGF stimulation condition (d). Immunoblot analysis (IB) of whole cell lysates (WCL) and Flag immunoprecipitates (IP) derived from HeLa cells transfected with Flag–Sin1 that were serum starved for 12 hours and then treated with insulin (100 nM) (c) or EGF (100 ng/ml) (d) for the indicated time periods before harvesting for IB analysis. e–f. Quantification curves for the relative pT86-Sin1 intensity and Rictor abundance as a function to the treatment time as presented in Supplementary Fig. 4c,d. g. Rapamycin or S6K1-I treatment led to a relatively sustained Akt activation upon EGF stimulation. IB analysis of WCL derived from Sin1 WT MEFs stimulated with EGF with indicated inhibitors. MEFs were serum starved for 12 hours and stimulated by 100 ng/ml EGF. Where indicated, cells were treated with 20 nM rapamycin or 10 μM S6K inhibitor (S6K1-I) for 2 hours before adding EGF and collecting the WCL at the indicated time points. h–i. Upon insulin or EGF stimulation, depletion of endogenous Raptor led to an elevated Akt-Ser473 phosphorylation while depletion of endogenous TSC2 caused a reduction in Akt-Ser473 phosphorylation. IB analysis of WCL derived from shGFP- or shRaptor- (h,i) or shTSC2 (i,j). HeLa cells that were serum starved for 24 hours and then treated with insulin (h and j) or EGF (i) for the indicated time periods before harvesting for IB analysis. k. Deletion of endogenous TSC2 led to attenuated Akt-Ser473 phosphorylation induced by insulin. IB analysis of WCL derived from TSC2+/+ or TSC2−/− MFEs that were serum starved for 18 hours and then treated with insulin for the indicated time periods before harvesting for IB analysis. l. Deletion of endogenous TSC2 led to attenuated Akt-Ser473 phosphorylation in response to insulin, IGF-1, PDGF or EGF stimulation. IB analysis of WCL derived from TSC2+/+ or TSC2−/− MFEs that were serum starved for 12 hours and then treated with the indicated stimuli for the indicated time periods before harvesting for IB analysis. Specifically, the doses of stimuli used are listed below: insulin (100 nM), IGF-1 (100 ng/ml), PDGF (100 ng/ml) and EGF (100 ng/ml). Other than the indicated time points for EGF, cells were treated for 30 min with other three stimuli before harvesting. m. Sin1 phospho-mimetic mutant (Sin1-EE) exhibited similar half-life with Sin1-WT. IB analysis of WCL derived from HeLa cells transfected with the indicated Flag–Sin1 constructs and harvested at the indicated time periods of CHX treatments (100 μg/ml). Mdm2 blots were provided as a control to indicate that CHX treatment was working. n. Depletion of endogenous TSC2, which resulted in elevated S6K kianse activity and subsequently increased Sin1 phosphorylation, did not lead to noticeable changes on the stability of endogenous Sin1. IB analysis of WCL derived from HeLa cells depleted of TSC2 via lentiviral infections (with shGFP as a negative control). Cells were harvested at the indicated time periods of CHX treatments (100 μg/ml). Mdm2 blots were provided as a control to indicate that CHX treatment was working.
Supplementary Figure 5 Sin1 phosphorylation could be tissue specific and might contribute to the timely shutting down of Akt phosphorylation triggered by insulin.
a-c. Insulin time course experiments performed in 3T3-L1 (a), HeLa (b) and OVCAR5 (c) cells indicate an oscillation pattern of Akt1-Ser473 and potential kinases responsible for both Sin1-T86 and Sin1-T398 phosphorylation. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from the indicated cells that were serum starved for 24 hours prior to insulin (100 nM) stimulation at the indicated time periods. Please note that the Sin1-pT398 signals were obtained by immunoblotting of the immunoprecipitated endogenous Sin1. d. A proposed model to illustrate a possible role of Sin1 phosphorylation in contributing to the oscillation of Akt activity upon the insulin stimulation. Specifically, at early time points, insulin initially triggers mTORC2 activation towards phosphorylating Akt, which subsequently activates mTORC1 to activate S6K. Afterwards, elevated S6K could directly phosphorylate Sin1 to disassemble mTORC2 to inactivate Akt in epithelial cells or fibroblasts. In adipocytes, Akt might be the major kinase phosphorylating Sin1. Thus, tissue or cell context-dependent phosphorylation of Sin1 might play a critical role in negatively regulating Akt phosphorylation in a timely fashion.
Supplementary Figure 6 Sin1 phosphorylation resulted in reduced Akt activity, subsequently sensitizing cells to etoposide-induced apoptosis.
a. Deletion of endogenous Sin1 led to dramatically reduced Akt-Ser473, but not Akt-Thr308, phosphorylation. WT or Sin1−/− MEFs were serum-starved for 24 hours and then collected after stimulation with the indicated stimuli for 30 minutes for immunoblot (IB) analysis. b-c. Cell viability assays with etoposide treatments to illustrate that etoposide triggers cellular apoptosis in part via the Akt/FOXO signaling pathway. HeLa cells depleted of endogenous Akt1 (b) or transfected with the indicated Flag-FOXO3a constructs (either Flag-FOXO3a-WT or Flag-FOXO3a-T32A, with empty vector as a negative control) (c) were cultured in 10% FBS-containing medium with the indicated concentrations of etoposide for 48 hours before performing the cell viability assays. Data was shown as mean ± SD from n = 3 independent experiments. d–e. Sin1-WT, but not Sin1-EE, could rescue the apoptotic deficiency observed in Sin1-depleted cells. IB analysis of WCL derived from endogenous Sin1-depleted OVCAR5 cells transfected with the indicated Flag–Sin1 constructs. Where indicated, various concentrations of etoposide were added 36 hours prior to cell collection. f–i. FACS analyses were performed to indicate that compared to shGFP-infected OVCAR cells (f), in endogenous Sin1-depleted OVCAR5 cells, stable expression of Sin1-WT (h), but not empty vector control (g) or Sin1-EE (i), could efficiently rescue the cellular apoptosis response triggered by etoposide (5 μM) for 24 hours. Data were presented as 7-AAD staining as a function to AnnexinV-PE staining. Cell populations were gated to illustrate the changes of the apoptotic cell population. Q1: dead cells; Q2: late apoptotic cells; Q3: non-apoptotic cells; Q4: early apoptotic cells. P1: apoptotic cells, equals to Q2+Q4.
Supplementary Figure 7 Cancer patient-derived Sin1 mutations led to an elevated and sustained Akt phosphorylation.
a–b. Like the Sin1-R81T mutation, Sin1-S84L led to reduced Sin1-pT86 phosphorylation in vivo. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from HeLa cells transfected with indicated HA-Sin1 constructs (a) or CMV-GST-Sin1-N-terminal-200aa constructs (b). c. The Sin1-R81T mutation stabilized Sin1 interaction with Rictor/mTOR upon insulin stimulation. IB analysis of WCL derived from Flag–Sin1-R81T-transfected HeLa cells that were serum-starved for 24 hours and then collected after insulin stimulation for the indicated time periods for Flag-IP. d. Quantification curves for the relative pT86-Sin1 intensity and Rictor abundance as a function to the treatment time as presented in Fig. 7e. e. Quantification curves for the relative pS473-Akt intensity in Sin1-WT or Sin1-R81T expressing OVCAR5 cells depleted of endogenous Sin1, as a function to the treatment time as presented in Fig. 7f. f. The Sin1-R81T mutation led to a relatively sustained Akt activation under PDGF stimulation. OVCAR5 ovarian cancer cells were depleted of endogenous Sin1 by shRNA constructs followed by re-introduction of Sin1-WT or the Sin1-R81T mutant via infection with the MSCV-retroviral vectors. The resulting cells were serum starved overnight followed by the addition of 100 ng/ml PDGF. Cells were harvested for IB analysis at the indicated time periods. g. A proposed model to describe how the ovarian cancer-derived Sin1-R81T mutation might protect the mTORC2 kinase from the negative regulation by phosphorylation of Sin1 on both the T86 and T398 sites. Specifically, the Sin1 R81T mutation led to elevated Akt S473 phosphorylation and subsequent Akt activation by bypassing the Sin1 phosphorylation mediated negative regulation of mTORC2/Akt signaling.
Supplementary Figure 8 The Sin1-R81T mutation led to cellular resistance to chemotherapeutic drugs and facilitated tumorigenesis.
a. Sin1-R81T led to elevated Akt Ser473 phosphorylation and subsequently, an increase in FOXO phosphorylation. Sin1−/− MEFs were transfected with the indicated Flag–Sin1 constructs (with empty vector as a negative control). 24 hours post-transfection, the resulting cells were serum starved and harvested after stimulation with 100 nM insulin for 30 minutes for immunoblot (IB) analysis b. Sin1-R81T led to elevated Akt Ser473 phosphorylation and subsequently, an increase in FOXO phosphorylation. OVCAR5 ovarian cancer cells were infected with the shSin1 (with shGFP as a negative control) lentiviral shRNA construct and selected with 250 μg/ml hygromycin for 72 hours to eliminate non-infected cells. Afterwards, the resulting cell lines were transfected with the indicated Flag–Sin1 constructs. 24 hours post-transfection, cells were serum-starved for 24 hours and then collected after stimulation with insulin for 30 minutes for IB analysis. c–d. Sin1 R81T restored cellular apoptotic deficiency in Sin1-deficient cells. IB analysis of whole cell lysates (WCL) derived from endogenous Sin1 depleted OVCAR5 cells transfected with the indicated Flag–Sin1 constructs. Where indicated, various concentrations of etoposide were added 36 hours prior to cell collection. e. Expression of Sin1-R81T conferred cellular resistance to etoposide. OVCAR5 ovarian cancer cells were infected with the shSin1 (with shGFP as a negative control) lentiviral construct and selected with 250 μg/ml hygromycin for 72 hours to eliminate non-infected cells. Afterwards, the resulting cells were infected with indicated MSCV-Sin1-Myc retroviral constructs (with empty vector as a negative control) and selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells. Obtained cell lines were cultured in 10% FBS-containing medium with indicated concentrations of etoposide for 48 hours before performing the cell viability assays. Data was shown as mean ±SD from n = 3 independent experiments. f-g. Sin1-R81T conferred cellular resistance to etoposide. Sin1−/− MEFs were transfected with the indicated Flag–Sin1 constructs and cultured in 10% FBS-containing medium with the indicated concentrations of etoposide for 24 hours (f) or 48 hours (g) before performing the cell viability assays. Data was shown as mean ±SD from n = 3 independent experiments. h. Representative images of tumors in vivo from the xenograft experiments presented in Fig. 8e-g. i. Soft agar assays to determine the critical role of the Akt oncogenic signaling in cellular transformation. Sub-confluent OVCAR5 cell lines were harvested for IB analysis to detect Akt expression. The above cell lines were also applied to soft agar assays with the presence of 4 μM AktVIII. Briefly, 1×105 cells were plated in the top layer containing 0.4% agar and 4 μM AktVIII. 3 weeks later cells were stained with iodonitrotetrazolium chloride for colony visualization and counting. n = 3 independent experiments were performed to generate the error bar, and data were presented as Mean ± SD. The scale bar represents 1 mm. j. Soft agar assays with the indicated OVCAR5 cell lines stably expressing Sin1-WT or Sin1-R81T mutant to determine their differential sensitivities to the Akt inhibitor, AktVIII treatment. The above cell lines were then applied to soft agar assay. Briefly, 3×105 cells were plated in the top layer containing 0.4% agar and the indicated amount of AktVIII. 3 weeks later cells were stained with iodonitrotetrazolium chloride for colony visualization and counting. n = 3 independent experiments were performed to generate the error bar, and data were presented as mean ± SD. The scale bar represents 1 mm. k. Cell viability assays to illustrate that the Sin1-R81T mutation led to acquired cellular resistance to AktVIII treatment. OVCAR5 cell lines stably expressing Sin1-WT or Sin1-R81T mutant were cultured in 10% FBS-containing medium with the indicated concentrations of AktVIII for 48 hours before performing the cell viability assays. Data was shown as mean ± SD from n = 3 independent experiments. l-o. Splenic B cells were enriched by negative selection from wild type B6 mice (l). Purified B cells were pretreated for 15 minutes with rapamycin (20 nM) then stimulated with anti-IgM antibody for an additional 30 minutes (m). Cells were lysed in CHAPS buffer, subjected to endogenous Sin1 immunoprecipitation and blotted with the indicated antibodies (m). Intensities of Sin1-pT86 bands (n) and Akt-pS473 bands (o) were quantified and normalized from two independent experiments. p. IB analysis of WCL from a panel of T-ALL cell lines. q. Two representative images of IHC results with indicated Sin1 and Akt phosphorylation status out of 58 ovarian patient samples under 400x magnification. The scale bar represents 100 μm. r. Bar graph of IHC statistical analysis using percentage of samples as a function to the relative levels of Sin1-T86 phosphorylation. 58 ovarian patient samples were analyzed by staining with either Akt-pS473 or Sin1-pT86 antibodies. p value <0.3 with Fisher’s exact analysis. s. A proposed model showing how phosphorylation of Sin1 by mTORC1/S6K or Akt in epithelial cells or adipocytes, respectively, actively suppresses the mTORC2/Akt activation in part by dissociating the phosphorylated form of Sin1 from the functional mTORC2 complex. Importantly, distinct from the mTORC1-mediated negative feedback loops involving either IRS-1 or Grb10 to suppress the PI3K/Akt pathway that only responds to insulin or IGF-1 stimulation, S6K-mediated phosphorylation of Sin1 in fibroblasts or epithelial cells, or Akt-mediated phosphorylation of Sin1 in adipocytes, can occur in response to multiple other stimuli including PDGF and EGF. Thus our work discovers an independent mode of negative regulation of mTORC2, ensuring no hyper-activation of the Akt oncoprotein that might facilitate tumorigenesis.
Supplementary information
Supplementary Information
Supplementary Information (PDF 3304 kb)
Rights and permissions
About this article
Cite this article
Liu, P., Gan, W., Inuzuka, H. et al. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat Cell Biol 15, 1340–1350 (2013). https://doi.org/10.1038/ncb2860
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2860
This article is cited by
-
AMPK-dependent phosphorylation of the GATOR2 component WDR24 suppresses glucose-mediated mTORC1 activation
Nature Metabolism (2023)
-
Aurora kinase a promotes the progression of papillary thyroid carcinoma by activating the mTORC2-AKT signalling pathway
Cell & Bioscience (2022)
-
Mechanistic target of rapamycin complex 1 orchestrates the interplay between hepatocytes and Kupffer cells to determine the outcome of immune-mediated hepatitis
Cell Death & Disease (2022)
-
Raptor downregulation rescues neuronal phenotypes in mouse models of Tuberous Sclerosis Complex
Nature Communications (2022)
-
Beyond controlling cell size: functional analyses of S6K in tumorigenesis
Cell Death & Disease (2022)