Abstract
Through in vivo selection of multiple ER-negative human breast cancer populations for enhanced tumour-forming capacity, we have derived subpopulations that generate tumours more efficiently than their parental populations at low cell numbers. Tumorigenic-enriched subpopulations exhibited increased expression of LAMA4, FOXQ1 and NAP1L3—genes that are also expressed at greater levels by independently derived metastatic subpopulations. These genes promote metastatic efficiency. FOXQ1 promotes LAMA4 expression, and LAMA4 enhances clonal expansion following substratum detachment in vitro, tumour re-initiation in multiple organs, and disseminated metastatic cell proliferation and colonization. The promotion of cancer cell proliferation and tumour re-initiation by LAMA4 requires β1-integrin. Increased LAMA4 expression marks the transition of human pre-malignant breast lesions to malignant carcinomas, and tumoral LAMA4 overexpression predicts reduced relapse-free survival in ER-negative patients. Our findings reveal common features that govern primary and metastatic tumour re-initiation and identify a key molecular determinant of these processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Klein, C. A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 9, 302–312 (2009).
Maheswaran, S. & Haber, D. A. Circulating tumor cells: a window into cancer biology and metastasis. Curr. Opin. Genet. Dev. 20, 96–99 (2010).
Chiang, A. C. & Massague, J. Molecular basis of metastasis. N. Engl. J. Med. 359, 2814–2823 (2008).
Fidler, I. J. The pathogenesis of cancer metastasis: the’ seed and soil ’hypothesis revisited. Nature Rev. Cancer 3, 453–458 (2003).
Pavlova, N. N. et al. A role for PVRL4-driven cell–cell interactions in tumorigenesis. eLife 2, e00358 (2013).
Williams, S. A., Anderson, W. C., Santaguida, M. T. & Dylla, S. J. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab. Invest. 93, 970–982 (2013).
Shimono, Y. et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 (2009).
Visvader, J. E. & Lindeman, G. J. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 10, 717–728 (2012).
Chaffer, C. L. et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61–74 (2013).
Magee, J. A., Piskounova, E. & Morrison, S. J. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21, 283–296 (2012).
Vanharanta, S. & Massague, J. Origins of metastatic traits. Cancer Cell 24, 410–421 (2013).
Clark, E. A., Golub, T. R., Lander, E. S. & Hynes, R. O. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406, 532–535 (2000).
Kang, Y. et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 (2003).
Minn, A. J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005).
Pencheva, N. et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell 151, 1068–1082 (2012).
Tavazoie, S. F. et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 (2008).
Png, K. J., Halberg, N., Yoshida, M. & Tavazoie, S. F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 481, 190–194 (2012).
Putti, T. C. et al. Estrogen receptor-negative breast carcinomas: a review of morphology and immunophenotypical analysis. Mod. Pathol. 18, 26–35 (2005).
Cailleau, R., Olive, M. & Cruciger, Q. V. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In vitro 14, 911–915 (1978).
Bos, P. D. et al. Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 (2009).
Rhim, A. D. et al. EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361 (2012).
Diehn, M. & Majeti, R. Metastatic cancer stem cells: an opportunity for improving cancer treatment? Cell Stem Cell 6, 502–503 (2010).
Brabletz, T., Jung, A., Spaderna, S., Hlubek, F. & Kirchner, T. Opinion: migrating cancer stem cells—an integrated concept of malignant tumour progression. Nat. Rev. Cancer 5, 744–749 (2005).
Brabletz, T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell 22, 699–701 (2012).
Padua, D. et al. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 133, 66–77 (2008).
Reymond, N., d’Agua, B. B. & Ridley, A. J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer 13, 858–870 (2013).
Nguyen, D. X., Bos, P. D. & Massague, J. Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer 9, 274–284 (2009).
Stenzel, D. et al. Endothelial basement membrane limits tip cell formation by inducing Dll4/Notch signalling in vivo. EMBO Rep. 12, 1135–1143 (2011).
Goering, W. et al. Impairment of gastric acid secretion and increase of embryonic lethality in Foxq1-deficient mice. Cytogenet. Genome Res. 121, 88–95 (2008).
Zhang, H. et al. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 71, 1292–1301 (2011).
Attia, M. et al. Interaction between nucleosome assembly protein 1-like family members. J. Mol. Biol. 407, 647–660 (2011).
Buchstaller, J., McKeever, P. E. & Morrison, S. J. Tumorigenic cells are common in mouse MPNSTs but their frequency depends upon tumor genotype and assay conditions. Cancer Cell 21, 240–252 (2012).
Grassian, A. R., Coloff, J. L. & Brugge, J. S. Extracellular matrix regulation of metabolism and implications for tumorigenesis. Cold Spring Harb. Symp. Quant. Biol. 76, 313–324 (2011).
Aguirre-Ghiso, J. A. Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer 7, 834–846 (2007).
Polyak, K. et al. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes Dev. 8, 9–22 (1994).
Zhu, X., Ohtsubo, M., Bohmer, R. M., Roberts, J. M. & Assoian, R. K. Adhesion-dependent cell cycle progression linked to the expression of cyclin D1, activation of cyclin E-cdk2, and phosphorylation of the retinoblastoma protein. J. Cell Biol. 133, 391–403 (1996).
Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K. & Elledge, S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 (1993).
Geberhiwot, T. et al. Laminin-8 (α4β1γ1) is synthesized by lymphoid cells, promotes lymphocyte migration and costimulates T cell proliferation. J. Cell Sci. 114, 423–433 (2001).
Fujiwara, H., Kikkawa, Y., Sanzen, N. & Sekiguchi, K. Purification and characterization of human laminin-8. Laminin-8 stimulates cell adhesion and migration through α3β1 and α6β1 integrins. J. Biol. Chem. 276, 17550–17558 (2001).
Kortesmaa, J., Yurchenco, P. & Tryggvason, K. Recombinant laminin-8 (α(4)β(1)γ(1)). Production, purification, and interactions with integrins. J. Biol. Chem. 275, 14853–14859 (2000).
Hohenester, E. & Yurchenco, P. D. Laminins in basement membrane assembly. Cell Adhes. Migration 7, 56–63 (2013).
Moreno-Layseca, P. & Streuli, C. H. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 34, 144–153 (2014).
Miranti, C. K. & Brugge, J. S. Sensing the environment: a historical perspective on integrin signal transduction. Nat. Cell Biol. 4, E83–90 (2002).
Giancotti, F. G. Mechanisms governing metastatic dormancy and reactivation. Cell 155, 750–764 (2013).
Shibue, T. & Weinberg, R. A. Integrin β1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc. Natl Acad. Sci. USA 106, 10290–10295 (2009).
Espina, V. & Liotta, L. A. What is the malignant nature of human ductal carcinoma in situ? Nat. Rev. Cancer 11, 68–75 (2011).
Lee, S. et al. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 72, 4574–4586 (2012).
Ma, X. J. et al. Gene expression profiles of human breast cancer progression. Proc. Natl Acad. Sci. USA 100, 5974–5979 (2003).
Ma, X. J., Dahiya, S., Richardson, E., Erlander, M. & Sgroi, D. C. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 11, R7 (2009).
Schuetz, C. S. et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 66, 5278–5286 (2006).
Desmedt, C. et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 13, 3207–3214 (2007).
Hatzis, C. et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA 305, 1873–1881 (2011).
van de Vijver, M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002).
Wang, Y. et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 (2005).
Peduto, L. et al. ADAM12 is highly expressed in carcinoma-associated stroma and is required for mouse prostate tumor progression. Oncogene 25, 5462–5466 (2006).
Kessenbrock, K., Plaks, V. & Werb, Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67 (2010).
Goss, P. E. & Chambers, A. F. Does tumour dormancy offer a therapeutic target? Nat. Rev. Cancer 10, 871–877 (2010).
Nelson, C. M. & Bissell, M. J. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 22, 287–309 (2006).
Kusuma, N. et al. Integrin-dependent response to laminin-511 regulates breast tumor cell invasion and metastasis. Int. J. Cancer 130, 555–566 (2012).
Yurchenco, P. D. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 3, a004911 (2011).
Fujita, M. et al. Overexpression of β1-chain-containing laminins in capillary basement membranes of human breast cancer and its metastases. Breast Cancer Res. 7, R411–421 (2005).
Hamill, K. J., Kligys, K., Hopkinson, S. B. & Jones, J. C. Laminin deposition in the extracellular matrix: a complex picture emerges. J. Cell Sci. 122, 4409–4417 (2009).
Ilani, T. et al. A secreted disulfide catalyst controls extracellular matrix composition and function. Science 341, 74–76 (2013).
Vainionpaa, N., Lehto, V. P., Tryggvason, K. & Virtanen, I. α4 chain laminins are widely expressed in renal cell carcinomas and have a de-adhesive function. Lab. Invest. 87, 780–791 (2007).
Jemal, A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011).
Ali, S. & Coombes, R. C. Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer 2, 101–112 (2002).
Ponomarev, V. et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur. J. Nucl. Med. Mol. Imaging 31, 740–751 (2004).
Hickson, J. et al. Noninvasive molecular imaging of apoptosis in vivo using a modified firefly luciferase substrate, Z-DEVD-aminoluciferin. Cell Death Differ. 17, 1003–1010 (2010).
Gazdar, A. F. et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int. J. Cancer. 78, 766–774 (1998).
Li, Q., Birkbak, N. J., Gyorffy, B., Szallasi, Z. & Eklund, A. C. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics 12, 474 (2011).
Acknowledgements
We are grateful to members of our laboratory for insightful discussion and C. Alarcon, N. Halberg, N. Pencheva and A. Nguyen for providing comments on previous versions of this manuscript. We thank C. Blobel, S. Simon and J. Friedman for intellectual input and helpful suggestions. We thank A. Nguyen and J. M. Loo for assistance with splenic injections. We thank N. Halberg for assistance with tail-vein injections. We thank H. Goodarzi for assistance with statistical analysis. We thank P. Furlow for cloning assistance. We thank C. Zhao of the Rockefeller Genomics Resource Center for assistance with transcriptomic profiling. We thank S. Mazel and members of the Rockefeller Flow Cytometry Resource Center for FACS sorting. J.B.R., D.H. and L.B.N. are members of the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program supported by NIH MSTP grant GM07739. S.F.T. is a DOD Era of Hope Scholar and Collaborative Scholars and Innovators Award recipient and Head of the Elizabeth and Vincent Meyer Laboratory of Systems Cancer Biology.
Author information
Authors and Affiliations
Contributions
S.F.T. conceived the project and supervised all research. J.B.R. and S.F.T. wrote the manuscript. J.B.R. conducted in vivo selection for tumour re-initiation. J.B.R., D.H. and L.B.N. designed, performed and analysed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 5 Characterization of tumorigenic-enriched derivatives.
(a) 2.5 × 104 MDA-parental or MDA-TE3 cells were seeded into 6-well adherent tissue-culture plates. The number of cells per well on day 5 was counted and normalized to cell counts on day 1. n = 3 independent wells. (b) 2.5 × 104 CN34-parental or CN34-TE2 cells were seeded into 6-well adherent tissue-culture plates. The number of cells per well on day 5 was counted and normalized to cell counts on day 1. n = 3 independent wells. (c) 1 × 102 MDA-parental or MDA-TE3 cells were seeded into 10cm adherent tissue-culture plates. The number of colonies per well on day 14 was counted upon staining with crystal-violet. n = 3 independent plates. (d) 1 × 102 CN34-parental or CN34-TE2 cells were seeded into 10cm adherent tissue-culture plates. The number of colonies per well on day 14 was counted upon staining with crystal-violet. n = 3 independent plates. (e) Assessment of cell attachment to adherent tissue culture plates of MDA-parental cells compared to MDA-TE3 cells. n = 3 independent wells. (f) Assessment of cell attachment to adherent tissue culture plates of CN34-parental cells compared to CN34-TE3 cells. n = 6 independent wells. (g) Endothelial recruitment assay comparing the relative capacity of MDA-parental cells and MDA-TE3 cells recruit Human Vein Endothelial Cells (HUVECs). n = 6 independent trans-well inserts. (h) Endothelial recruitment assay comparing the relative capacity of CN34-parental cells and CN34-TE3 cells recruit Human Vein Endothelial Cells (HUVECs). n = 8 independent trans-well inserts. (i) MDA-parental and MDA-TE3 populations were stained with antibodies specific to the cell surface markers CD44 and CD24 and analyzed using flow cytometry. For the MDA-parental population, 86.1% of the cells were CD44 + CD24-. For the MDA-TE3 population, 84.4% of the cells were CD44 + CD24-. (j) CN34-parental and CN34-TE2 populations were stained with antibodies specific to the cell surface markers CD44 and CD24 and analyzed using flow cytometry. For the CN34-parental population, 89.1% of the cells were CD44 + CD24-. For the CN34-TE2 population, 86.1% of the cells were CD44 + CD24-. NS is not significant. ∗∗P < 0.01,∗∗∗P < 0.001 were obtained using a two-sided student’s t-test (a–h). All data are represented as mean ± S.E.M.
Supplementary Figure 6 Independent shRNA knockdown of LAMA4, FOXQ1, and NAP1L3 leads to suppression of metastasis in vivo.
(a) 1.5 × 105 CN34-LM1a cells transduced with either a control shRNA hairpin or an independent shRNA hairpin targeting LAMA4 were inoculated intravenously into immunodeficient mice. shRNA depletion of LAMA4 led to a significant reduction in metastasis as measured by bioluminescence imaging on day 25 normalized to post-injection signal on day 0. n = 4 (shControl), n = 3 (shLAMA4_2) independent mice. (b) 5 × 104 CN34-LM1a cells transduced with either a control shRNA hairpin or an independent shRNA hairpin targeting FOXQ1 were inoculated intravenously into immunodeficient mice. shRNA depletion of FOXQ1 led to a significant reduction in metastasis as measured by bioluminescence imaging on day 84 normalized to post-injection signal on day 0. n = 4 (shControl), n = 5 (shFOXQ1_2) independent mice. (c) 5 × 104 CN34-LM1a cells transduced with either a control shRNA hairpin or an independent shRNA hairpin targeting NAP1L3 were inoculated intravenously into immunodeficient mice. shRNA depletion of NAP1L3 led to a significant reduction in metastasis as measured by bioluminescence imaging on day 42 normalized to post-injection signal on day 0. n = 4 independent mice. ∗P < 0.05,∗∗P < 0.01, were obtained using one-sided Mann–Whitney test (a–c). All data are represented as mean + S.E.M.
Supplementary Figure 7 Assessment of laminin-α4 protein levels in vitro and in vivo.
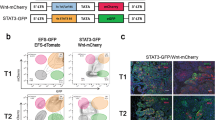
(a) Anti-laminin-α4 antibody was used to detect endogenous LAMA4 protein in supernatant collected from in vitro culture of MDA-parental, MDA-TE3, and MDA-LM2 cells. (b) Anti-laminin-α4 antibody was used to detect endogenous LAMA4 protein in supernatant collected from in vitro culture of CN34-parental and CN34-TE2 cells. (c) Anti-laminin-α4 antibody was used to detect endogenous protein in supernatant collected from in vitro culture of MDA-parental empty vector control (C), MDA-parental LAMA4-overexpression (oe), MDA-TE3 shControl (shC), MDA-TE3 shLAMA4_1 (sh1), and MDA-TE3 shLAMA4_2 (sh2) cells. All Western blots (a–c) are representative and based on multiple experiments (see Supplementary Fig. 8 for un-cropped images). (d) Anti-human laminin-α4 antibody was used to detect endogenous LAMA4 protein in xenograft tumors derived from MDA-parental, MDA-TE3, or MDA-LM2 populations. Upper panels: Red is anti-laminin-α4 antibody (LAMA4), Blue is DAPI. Core is representative of tumor core, Edge is representative of tumor edge. Lower panels: Red is non-specific isotype-matched control antibody (IgG), Blue is DAPI. Core is representative of tumor core. Scale bars: 25 μm (d) Immunofluorescence of xenograft tumors at higher magnification. Co-staining of xenograft tumors with anti-laminin-α4 antibody and anti-Collagen-IV antibody revealed areas of overlap. Red is anti-laminin-α4 antibody (LAMA4), Green is anti-Collagen-IV antibody (Col-IV), Blue is DAPI. Scale bars: 25 μm. Immunofluorescence (d–e) is representative of multiple samples/experiments.
Supplementary Figure 8 LAMA4 is sufficient to promote metastatic colonization and promotes tumor re-initiation from low cell numbers.
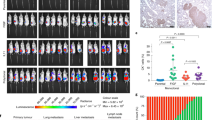
(a–b) 2 × 105 HCC1806 cells transfected with either a control siRNA or two independent siRNAs targeting LAMA4 were inoculated intravenously into immunodeficient mice. siRNA depletion of LAMA4 led to a significant reduction in metastasis as measured by bioluminescence imaging on day 42 normalized to post-injection signal on day 0 (a). n = 6 independent mice. Lungs were harvested on day 42, H&E-stained, and the number of macroscopic nodules per lung section was counted. Representative lungs on day 42 (b). n = 6 (siControl), n = 5 (siLAMA4_5), n = 6 (siLAMA4_6) lungs from independent mice. Scale bars: 1mm. Insets are magnified 5×. (c–d) 5 × 105 CN34-parental cells transduced with either an empty vector control or LAMA4 over-expression vector were inoculated intravenously into immunodeficient mice. Lung bioluminescence was measured on day 133 and normalized to post-injection signal at day 0 (c). n = 7 independent mice. Lungs were harvested on day 133, vimentin-stained, and the number of macroscopic nodules per lung section was counted. Representative vimentin-stained lungs on day 133 (d). n = 7 lungs from 7 independent mice. Scale bars: 1mm. Insets are magnified 5×. (e) 1 × 101 MDA-TE3 cells transduced with either a control shRNA or an independent shRNA targeting LAMA4 were injected into the mammary fat pads of immunodeficient mice. MDA-TE3 shControl cells yielded tumors in 14/24 sites as compared to 6/24 sites for MDA-TE3 shLAMA4_2 after 10 weeks (left). n = 24 independent mammary fat pad injections (pooled from 6 mice with 4 injections each per condition and represented as open squares, right). (f) The number of tumors formed upon injection of 5 × 105 MDA-TE3 cells transduced with either a control shRNA or two independent shRNAs targeting LAMA4 into the mammary fat pads of immunodeficient mice was quantified (see Fig. 4f). Tumors were formed in all cases. n = 8 independent mammary fat pad injections (pooled from 2 mice with 4 injections each per condition). (g,h) 1 × 102 MDA-TE3 cells transduced with either a control shRNA or an independent shRNA targeting LAMA4 were injected directly into the lung parenchyma to assess ectopic tumor re-initiation capacity. Lung bioluminescence was measured on day 63 (g). n = 5 independent mice. On day 63 lungs were sectioned, vimentin stained, and the number of macroscopic nodules per lung was counted (h). n = 5 lungs from 5 independent mice. Scale bars: 1mm. Insets are magnified 5×.∗P < 0.05,∗∗P < 0.01 were obtained using one-sided Mann–Whitney test (a–d,g–h) or one-sided Fisher’s exact test (e). All data are represented as mean + S.E.M.
Supplementary Figure 9 LAMA4’s promotion of proliferation in vitro and tumor re-initiation in vivo requires β1-integrin.
(a) Cells were sorted at clonal density (one cell per well) into low-attachment 96-well plates to assess proliferation in the absence of substratum-attachment. (b) MDA-parental, MDA-TE3, or MDA-LM2 cells were sorted at clonal density into low-attachment plates. The number of wells containing one cell versus the number of wells containing multiple cells was assessed on D3. n = 145 (parental), n = 156 (TE3), n = 133 (LM2) wells. Data from Fig. 5b. (c) MDA-TE3-control cells or MDA-TE3-LAMA4-knockdown cells were sorted at clonal density into low-attachment plates. The number of wells containing one cell compa versus the number of wells containing multiple cells was assessed on D3. n = 189 (shControl), n = 196 (shLAMA4_1), n = 197 (shLAMA4_2) wells. Data from Fig. 5a. (d) Cells were seeded at low densitiy into low-attachment 6-well plates in high-viscosity media and isolated for cell-cycle analysis. (e) Cell-cycle analysis of MDA-TE3-control cells or MDA-TE3-LAMA4-knockdown cells. n = 5 samples. (f) Ki67-positive fraction was assessed in MDA-TE3-control cells or MDA-TE3-LAMA4-knockdown cells. n = 6 samples. (g) β1-integrin blocking antibody suppressed the proliferation of CN34-TE2 cells upon sorting at clonal density into low-attachment plates. Counts on D3. n = 383 (IgG), n = 438 (β1) wells. (h) The reduced proliferation of MDA-TE3-LAMA4-knockdown cells was rescued by recombinant LAMA4-containing protein upon sorting at clonal density into low-attachment plates. Counts on D3. n = 4 independent experiments. (i) Incubation of MDA-Parental-control cells with β1-integrin blocking antibody did not lead to significant changes in proliferation upon sorting at clonal density into low-attachment plates (in contrast to MDA-Parental-LAMA4-overexpressing cells, Fig. 5g). Counts on D3. n = 233 (IgG), n = 221 (β1), n = 214 (αvβ3) wells. (j) Incubation of MDA-TE3-shControl cells with laminin-411 did not lead to significant changes in proliferation upon sorting at clonal density into low-attachment plates (in contrast to MDA-TE3-LAMA4-knockdown cells, Supplementary Fig. 5h). Counts on D3. n = 237 (neg), n = 242 (BSA), n = 269 (411) wells. (k) Pre-incubation of MDA-TE3-shControl cells with β1-integrin blocking antibody led to decreased proliferation independent of the addition of laminin-411 upon sorting at clonal density into low-attachment plates. Counts on D3. n = 169 (IgG + BSA), n = 337 (IgG + 411), n = 299 (β1 + BSA), n = 313 (β1 + 411) wells. (l) Western of lysates from MDA-TE3-control cells or MDA-TE3-LAMA4-knockdown cells cultured in the absence of substratum attachment. Western representative and based on multiple experiments (Supplementary Fig. 8, un-cropped images). ∗P < 0.05,∗∗P < 0.01,∗∗∗P < 0.001 were obtained using one-sided Fisher’s exact test (b,c), one-sided student’s t-test (e,f,h) or one-sided Mann–Whitney test (g,i–k). All data are represented as mean + S.E.M.
Supplementary Figure 10 LAMA4 does not significantly regulate apoptosis at early time points during metastatic colonization.
(a) 3 × 105 CN34-LM1a cells transduced with either control shRNA or an shRNA targeting LAMA4 were injected intravenously into immunodeficient mice (see Fig. 6a, b). In vivo quantification of apoptotic cells was monitored by measurement of a luciferase-based caspase-3/7 reporter normalized to cancer cell luciferase signal over several days (left). Representative luciferase-based caspase-3/7 (non-normalized) bioluminescence signal (right). n = 5 independent mice. NS is not significant based on a two-sided Student’s t-test. (b) Cumulative fraction plot depicting the distribution of metastatic foci size in the lungs of animals injected with CN34-LM1a cells that were transduced with either a control shRNA hairpin or an shRNA hairpin targeting LAMA4 (see Fig. 6a, b). n = 5 lungs from 5 independent mice. (c) Cumulative fraction plot depicting the distribution of metastatic foci size in the lungs of animals injected with CN34-LM1a cells that were transduced with either a control shRNA hairpin or two independent shRNA hairpins targeting LAMA4 (see Fig. 6e, f). n = 4 lungs from 4 independent mice. P values were obtained using a Kolmogorov-Smirnov test. All data are represented as mean ± S.E.M.
Supplementary Figure 11 Assessment of LAMA4 expression upon depletion of NAP1L3.
(a–b) Relative expression of LAMA4 and NAP1L3 following siRNA knockdown of NAP1L3 in LM1as (a; n = 12) or LM2s (b; n = 10) compared to control siRNA. n is the number of samples collected over 4 (a) or 3 (b) independent experiments. NS is not significant. ∗P < 0.05,∗∗∗P < 0.001 were obtained using a two-sided one-sample t-test (a–b). All data are represented as mean ± S.E.M.
Supplementary Figure 12 Un-cropped Western blots.
Rectangle includes cropped images present in Supplementary Fig. 3 (a–c) and Supplementary Fig. 5 (l).
Supplementary information
Supplementary Information
Supplementary Information (PDF 2278 kb)
Rights and permissions
About this article
Cite this article
Ross, J., Huh, D., Noble, L. et al. Identification of molecular determinants of primary and metastatic tumour re-initiation in breast cancer. Nat Cell Biol 17, 651–664 (2015). https://doi.org/10.1038/ncb3148
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3148
This article is cited by
-
FOXQ1 recruits the MLL complex to activate transcription of EMT and promote breast cancer metastasis
Nature Communications (2022)
-
Targeting the interaction between RNA-binding protein HuR and FOXQ1 suppresses breast cancer invasion and metastasis
Communications Biology (2020)
-
Gene expression analysis to detect disseminated tumor cells in the bone marrow of triple-negative breast cancer patients predicts metastatic relapse
Breast Cancer Research and Treatment (2019)
-
MiR-539 inhibits proliferation and migration of triple-negative breast cancer cells by down-regulating LAMA4 expression
Cancer Cell International (2018)
-
Cancer metastasis: enactment of the script for human reproductive drama
Cancer Cell International (2017)