Abstract
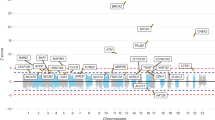
Ewing sarcoma, a pediatric tumor characterized by EWSR1-ETS fusions, is predominantly observed in populations of European ancestry. We performed a genome-wide association study (GWAS) of 401 French individuals with Ewing sarcoma, 684 unaffected French individuals and 3,668 unaffected individuals of European descent and living in the United States. We identified candidate risk loci at 1p36.22, 10q21 and 15q15. We replicated these loci in two independent sets of cases and controls. Joint analysis identified associations with rs9430161 (P = 1.4 × 10−20; odds ratio (OR) = 2.2) located 25 kb upstream of TARDBP, rs224278 (P = 4.0 × 10−17; OR = 1.7) located 5 kb upstream of EGR2 and, to a lesser extent, rs4924410 at 15q15 (P = 6.6 × 10−9; OR = 1.5). The major risk haplotypes were less prevalent in Africans, suggesting that these loci could contribute to geographical differences in Ewing sarcoma incidence. TARDBP shares structural similarities with EWSR1 and FUS, which encode RNA binding proteins, and EGR2 is a target gene of EWSR1-ETS. Variants at these loci were associated with expression levels of TARDBP, ADO (encoding cysteamine dioxygenase) and EGR2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Delattre, O. et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359, 162–165 (1992).
Aurias, A. et al. Chromosome translocation in Ewing's sarcoma. N. Engl. J. Med. 309, 496–498 (1983).
Toomey, E.C., Schiffman, J.D. & Lessnick, S.L. Recent advances in the molecular pathogenesis of Ewing's sarcoma. Oncogene 29, 4504–4516 (2010).
Linden, G. & Dunn, J.E. Ewing's sarcoma in Negroes. Lancet 295, 1171 (1970).
Jensen, R.D. & Drake, R.M. Rarity of Ewing's tumour in Negroes. Lancet 295, 777 (1970).
Li, F.P. et al. Rarity of Ewing's sarcoma in China. Lancet 315, 1255 (1980).
Jawad, M.U. et al. Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: an analysis of 1631 cases from the SEER database, 1973–2005. Cancer 115, 3526–3536 (2009).
Worch, J. et al. Racial differences in the incidence of mesenchymal tumors associated with EWSR1 translocation. Cancer Epidemiol. Biomarkers Prev. 20, 449–453 (2011).
Hutter, R.V., Francis, K.C. & Foote, F.W. Jr. Ewing's sarcoma in siblings: report of the second known occurrence. Am. J. Surg. 107, 598–603 (1964).
Joyce, M.J. et al. Ewing's sarcoma in female siblings. A clinical report and review of the literature. Cancer 53, 1959–1962 (1984).
Fraley, C. & Raftery, A.E. MCLUST version 3 for R: normal mixture modeling and model-based clustering (technical report no. 504) (Department of Statistics, University of Washington, Seattle, Washington, 2006).
Brisset, S. et al. CGH analysis of secondary genetic changes in Ewing tumors: correlation with metastatic disease in a series of 43 cases. Cancer Genet. Cytogenet. 130, 57–61 (2001).
Savola, S. et al. Combined use of expression and CGH arrays pinpoints novel candidate genes in Ewing sarcoma family of tumors. BMC Cancer 9, 17 (2009).
Clavel-Chapelon, F. et al. E3N, a French cohort study on cancer risk factors. E3N Group. Etude Epidémiologique auprès de femmes de l'Education Nationale. Eur. J. Cancer Prev. 6, 473–478 (1997).
Barmada, S.J. & Finkbeiner, S. Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: disease-associated pathways. Rev. Neurosci. 21, 251–272 (2010).
Chiang, P.M. et al. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc. Natl. Acad. Sci. USA 107, 16320–16324 (2010).
Lagier-Tourenne, C., Polymenidou, M. & Cleveland, D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64 (2010).
Dominy, J.E. et al. Discovery and characterization of a second mammalian thiol dioxygenase, cysteamine dioxygenase. J. Biol. Chem. 282, 25189–25198 (2007).
Schneider-Maunoury, S. et al. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell 75, 1199–1214 (1993).
Topilko, P. et al. Krox-20 controls myelination in the peripheral nervous system. Nature 371, 796–799 (1994).
Berger, P., Niemann, A. & Suter, U. Schwann cells and the pathogenesis of inherited motor and sensory neuropathies (Charcot-Marie-Tooth disease). Glia 54, 243–257 (2006).
Levi, G. et al. Defective bone formation in Krox-20 mutant mice. Development 122, 113–120 (1996).
Gabet, Y. et al. Krox20/EGR2 deficiency accelerates cell growth and differentiation in the monocytic lineage and decreases bone mass. Blood 116, 3964–3971 (2010).
Leclerc, N. et al. Opposing effects of glucocorticoids and Wnt signaling on Krox20 and mineral deposition in osteoblast cultures. J. Cell. Biochem. 103, 1938–1951 (2008).
Hancock, J.D. & Lessnick, S.L. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle 7, 250–256 (2008).
Tirode, F. et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 11, 421–429 (2007).
Chung, C.C. & Chanock, S.J. Current status of genome-wide association studies in cancer. Hum. Genet. 130, 59–78 (2011).
Maris, J.M. et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N. Engl. J. Med. 358, 2585–2593 (2008).
Johnson, A.D. et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24, 2938–2939 (2008).
Sladek, R. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 (2007).
Yeager, M. et al. Genome-wide association study of prostate cancer identifies a second locus at 8q24. Nat. Genet. 39, 645–649 (2007).
Hunter, D.J. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874 (2007).
Purcell, S. et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 81, 559–575 (2007).
Barrett, J.C., Fry, B., Maller, J. & Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Williamson, D. Fusion gene–negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J. Clin. Oncol. 28, 2151–2158 (2010).
Acknowledgements
We thank E. Thomas from Synergie Lyon Cancer for advice on statistical methods. We thank the staff from the Integragen Company for excellent service and V. Chene for outstanding assistance. We thank the following clinicians for providing samples used in this work: C. Alenda, F. Almazán, D. Ansoborlo, L. Aymerich, L. Benboukbher, C. Beléndez, C. Berger, C. Bergeron, P. Biron, J.Y. Blay, E. Bompas, H. Bonnefoi, P. Boutard, B. Bui-Nguyen, D. Chauveaux, C. Calvo, A. Carboné, C. Clement, T. Contra, N. Corradini, A.S. Defachelles, V. Gendemer-Delignieres, A. Deville, A. Echevarria, J. Fayette, M. Fraga, D. Frappaz, J.L. Fuster, P. García-Miguel, J.C. Gentet, P. Kerbrat, V. Laithier, V. Laurence, P. Leblond, O. Lejars, R. López-Almaraz, B. López-Ibor, P. Lutz, J.F. Mallet, L. Mansuy, P. Marec Bérard, G. Margueritte, A. Marie Cardine, C. Melero, L. Mignot, F. Millot, O. Minckes, G. Margueritte, C. Mata, M.E. Mateos, M. Melo, C. Moscardó, M. Munzer, B. Narciso, A. Navajas, D. Orbach, C. Oudot, H. Pacquement, C. Paillard, Y. Perel, T. Philip, C. Piguet, M.I. Pintor, D. Plantaz, E. Plouvier, S. Ramirez-Del-Villar, I. Ray-Coquard, Y. Reguerre, M. Rios, P. Rohrlich, H. Rubie, A. Sastre, G. Schleiermacher, C. Schmitt, P. Schneider, L. Sierrasesumaga, C. Soler, N. Sirvent, S. Taque, E. Thebaud, A. Thyss, R. Tichit, J.J. Uriz, J.P. Vannier, F. Watelle-Pichon. We also thank the European Prospective Investigation into Cancer and Nutrition (EPIC) Steering Committee (E. Riboli, Principal Investigator) for access to genotype data from the EPIC cohorts and the Children's Cancer and Leukaemia Group (CCLG) Tissue Bank (funded by Cancer Research UK) for access to specimens. This work was supported by grants from the Institut Curie, the Inserm, the Ligue Nationale Contre le Cancer (Equipe labellisée, Carte d'Identité des Tumeurs program and Recherche Epidémiologique 2009 program), the Région Ile de France, the Institut National du Cancer (2008-044, 0627 and ZP09-027-EPI), the Synergie Lyon Cancer foundation, the KCK and European Embryonal Tumor (EET) pipeline programs, the Société Française des Cancers de l'Enfant, the Spanish Ministry of Science and Technology (SAF2009-10158), the Fundación de la Asociación Española Contra el Cáncer, the Fundación María Francisca de Roviralta and the Sociedad Española de Hematología y Oncología Pediátricas and the following associations: Courir pour Mathieu, Dans les pas du Géant, Olivier Chape, Les Bagouzamanon, Enfants et Santé and les Amis de Claire.
Author information
Authors and Affiliations
Contributions
S.P.-V. contributed to the design of the study and wrote all documents required for ethical issues as well as information for patients and referent oncologists. A.S.V., D.C. and G.T. analyzed the GWAS data. G.P., S.B. and S.R. prepared all samples and performed genotyping of replication cohorts. F.T. and C.L. analyzed transcriptomic data and performed genotype-expression correlation analyses. H.K., O.O., U.K., A.G.-N., P.P., J.A., A.P.-G., B.B.d.P., F.D., J.M. and O.D. recruited Ewing sarcoma patients and participated in the diagnostic evaluations. E.L. collected informed consents. K.L. contributed the conditional Ewing cell model and performed the in vivo experiments. C.D., P.F. and F.C.-C. contributed French, E3N and EPIC controls. S.P.V., G.T., S.J.C., D.G.C. and O.D. contributed to the overall design of the study. All coauthors contributed to manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–4 and Supplementary Figures 1–6 (PDF 4201 kb)
Rights and permissions
About this article
Cite this article
Postel-Vinay, S., Véron, A., Tirode, F. et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet 44, 323–327 (2012). https://doi.org/10.1038/ng.1085
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.1085
This article is cited by
-
Identification of germline cancer predisposition variants in pediatric sarcoma patients from somatic tumor testing
Scientific Reports (2023)
-
Therapeutic targeting the oncogenic driver EWSR1::FLI1 in Ewing sarcoma through inhibition of the FACT complex
Oncogene (2023)
-
Integrative analyses of bulk microarray data to discover genes, pathways, and immune infiltration characteristics associated with targeting of Ewing sarcoma
Journal of Cancer Research and Clinical Oncology (2023)
-
Birth characteristics and risk of Ewing sarcoma
Cancer Causes & Control (2023)
-
TrkC, a novel prognostic marker, induces and maintains cell survival and metastatic dissemination of Ewing sarcoma by inhibiting EWSR1-FLI1 degradation
Cell Death & Disease (2022)