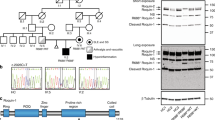
Abstract
Upon detection of pathogen-associated molecular patterns, innate immune receptors initiate inflammatory responses. These receptors include cytoplasmic NOD-like receptors (NLRs) whose stimulation recruits and proteolytically activates caspase-1 within the inflammasome, a multiprotein complex. Caspase-1 mediates the production of interleukin-1 family cytokines (IL1FCs), leading to fever and inflammatory cell death (pyroptosis)1,2. Mutations that constitutively activate these pathways underlie several autoinflammatory diseases with diverse clinical features3. We describe a family with a previously unreported syndrome featuring neonatal-onset enterocolitis, periodic fever, and fatal or near-fatal episodes of autoinflammation. We show that the disease is caused by a de novo gain-of-function mutation in NLRC4 encoding a p.Val341Ala substitution in the HD1 domain of the protein that cosegregates with disease. Mutant NLRC4 causes constitutive IL1FC production and macrophage cell death. Infected macrophages from affected individuals are polarized toward pyroptosis and exhibit abnormal staining for inflammasome components. These findings identify and describe the cause of a life-threatening but treatable autoinflammatory disease that underscores the divergent roles of the NLRC4 inflammasome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Mariathasan, S. & Monack, D.M. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat. Rev. Immunol. 7, 31–40 (2007).
Dinarello, C.A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27, 519–550 (2009).
Ozen, S. & Bilginer, Y. A clinical guide to autoinflammatory diseases: familial Mediterranean fever and next-of-kin. Nat. Rev. Rheumatol. 10, 135–147 (2014).
Thornberry, N.A. et al. A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature 356, 768–774 (1992).
Miao, E.A. et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7, 569–575 (2006).
Sutterwala, F.S. et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204, 3235–3245 (2007).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Mariathasan, S. et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 (2004).
Broz, P., von Moltke, J., Jones, J.W., Vance, R.E. & Monack, D.M. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8, 471–483 (2010).
Miao, E.A. et al. Caspase-1–induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11, 1136–1142 (2010).
Jordan, M.B., Allen, C.E., Weitzman, S., Filipovich, A.H. & McClain, K.L. How I treat hemophagocytic lymphohistiocytosis. Blood 118, 4041–4052 (2011).
Hu, Z. et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175 (2013).
Hoffman, H.M., Mueller, J.L., Broide, D.H., Wanderer, A.A. & Kolodner, R.D. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29, 301–305 (2001).
Agostini, L. et al. NALP3 forms an IL-1β–processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20, 319–325 (2004).
Aksentijevich, I. et al. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 46, 3340–3348 (2002).
Neven, B., Prieur, A.M. & Quartier dit Maire, P. Cryopyrinopathies: update on pathogenesis and treatment. Nat. Clin. Pract. Rheumatol. 4, 481–489 (2008).
Fernandes-Alnemri, T. et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 14, 1590–1604 (2007).
Man, S.M. et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA 111, 7403–7408 (2014).
Canna, S.W. et al. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 10.1038/ng.3089 (14 September 2014).
Franchi, L. et al. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 13, 449–456 (2012).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Lachmann, H.J. et al. Use of canakinumab in the cryopyrin-associated periodic syndrome. N. Engl. J. Med. 360, 2416–2425 (2009).
Hawkins, P.N., Lachmann, H.J., Aganna, E. & McDermott, M.F. Spectrum of clinical features in Muckle-Wells syndrome and response to anakinra. Arthritis Rheum. 50, 607–612 (2004).
Hoffman, H.M. et al. Efficacy and safety of rilonacept (interleukin-1 Trap) in patients with cryopyrin-associated periodic syndromes: results from two sequential placebo-controlled studies. Arthritis Rheum. 58, 2443–2452 (2008).
Bernstein, F.C. et al. The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur. J. Biochem. 80, 319–324 (1977).
Zaidi, S. et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223 (2013).
Vijayan, D. Isolation and differentiation of monocytes-macrophages from human blood. Methods Mol. Biol. 844, 183–187 (2012).
Wangdi, T., Mijares, L.A. & Kazmierczak, B.I. In vivo discrimination of type 3 secretion system–positive and –negative Pseudomonas aeruginosa via a caspase-1–dependent pathway. Infect. Immun. 78, 4744–4753 (2010).
Acknowledgements
We thank the patients and their families for their participation. We thank the Yale pediatric and medicine intensive care teams for providing access to patients. We thank the staff of the Yale Center for Genome Analysis for the generation of the exome sequence data and the Yale Center for Clinical Investigation for statistical assistance. We thank C. Roy (Yale School of Medicine) for the CASP1 and ASC constructs and L. Devine for multiplex cytokine detection. This work was supported by grants K12HD0141401-10 (US National Institutes of Health/National Institute of Child Health and Human Development) to N.R., U54HG006504 01 (Yale Center for Mendelian Genomics) to R.P.L. and R01AI081825 (US National Institutes of Health/National Institute of Allergy and Infectious Diseases) to B.I.K. B.I.K. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
Author information
Authors and Affiliations
Contributions
R.P.L., B.I.K., N.R. and K.A.M. designed the study and experiments. K.A.M. and N.R. performed cellular and molecular experiments. N.R., N.A.P., M.K.K. and E.L. identified, consented and recruited study subjects and provided clinical information. B.W. and A.J.H. provided autopsy and/or biopsy interpretations and images. C.N.-W., J.O. and M.C. generated and analyzed sequencing data. T.J.B. and A.L.S. generated and interpreted three-dimensional alteration maps for NLRC4. N.R. and E.M. processed case samples. N.R., R.P.L. and B.I.K. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
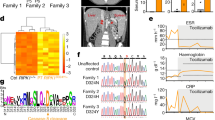
Supplementary Figure 1 The inflammatory cells infiltrating the central nervous system of deceased patient III.3 are mostly activated (CD163+) macrophages.
(a) Immunohistochemical staining of the patient’s choroid plexus shows the presence of numerous CD163+ macrophages. A vessel cut in cross-section reveals perivascular infiltration with CD163+ macrophages (inset). CD163+ macrophages also diffusely infiltrate the (b) leptomeninges and the (c) cortical perivascular space. Original magnification: 200× (for a and b), 100× (for c) and 400× (for the inset). Scale bars, 200 μm.
Supplementary Figure 2 SCAN4 macrophages show constitutive activation.
(a, right) A fraction of uninfected macrophages from SCAN4 patients II.3 and III.2 but not from a representative healthy donor (left) spontaneously formed ASC foci (red puncta; inlay) and stained positive for biotin-YVAD-CMK (green cytoplasmic staining). (b) Frequencies of macrophages positive for biotin-YVAD-CMK and ASC foci from SCAN4 patients (n = 2) and healthy controls (n = 2) visualized across multiple microscopic fields. Scale bars, 20 μm. Bar graphs show mean ± s.e.m. Significance by unpaired Student’s t test is indicated.
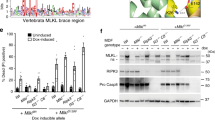
Supplementary Figure 3 In SCAN4 macrophages, constitutive IL-1 family cytokine secretion but not spontaneous cell death is dependent upon the caspase-1 catalytic site that processes cytokines.
(a) IL-1β and IL-18 secretion from monocyte-derived macrophages from SCAN4 patients (II.3 and III.2) cultured for 18 h in medium containing low-dose LPS (1 ng/ml) was attenuated by the caspase-1 inhibitor Z-YVAD-FMK (0.1 or 0.5 μM) relative to vehicle control (DMSO). (b) Cell death–associated LDH release by SCAN4 macrophages over the same 18-h period is not affected by Z-YVAD-FMK. LDH release is reported relative to cells lysed with Triton X-100 (0.1%). Bar graphs show mean supernatant values from experiments performed in duplicate or triplicate, and error bars represent ± s.e.m. Significance by paired Student’s t test is indicated.
Supplementary Figure 4 SCAN4 macrophages infected with pathogenic strains result in increased cell death yet blunted IL-1β and IL-18 production
(a) Monocytes were treated with LPS and infected with pathogenic strains PAKΔSTY and SL1344 or non-pathogenic strain PAKΔpopD as described in the Online Methods. (a) Secretion of IL-1β and IL-18 and (b) LDH release were measured in three independent experiments. Significant differences between subject groups assessed by unpaired Student’s t test have P values noted. Significant differences between wild-type cells infected with PAKΔSTY or SL1344 versus wild-type cells infected with PAKΔpopD assessed by paired Student’s t tests are noted by an asterisk when P < 0.05.
Supplementary Figure 5 SCAN4 macrophages infected with P. aeruginosa strain PAKΔSTY form increased numbers of ASC foci but show limited activation of caspase-1.
(a) The majority of PAKΔSTY-infected monocyte-derived macrophages from a representative healthy control (HD II.2; the mother of SCAN4 patient III.2) display features of conventional macrophage activation, including biotin-YVAD-CMK staining (green cytoplasmic staining) and a limited number (1–3) of ASC foci (red puncta) per cell (left). Many PAKΔSTY-infected macrophages (white arrows) from SCAN4 patients II.3 and III.2 but not from healthy controls display numerous (>6) ASC foci but show limited staining for biotin-YVAD-CMK (right). (b) Quantification of ASC foci in 472 PAKΔSTY-infected healthy control macrophages and 510 PAKΔSTY-infected patient macrophages. The patient-derived cell distribution was significantly different (P < 0.0001) from the healthy control distribution by χ2 testing. (c) Representative biotin-YVAD-CMK staining in PAKΔSTY-infected healthy control or SCAN4 macrophages from groups with either 1–3, 4–6 or >6 ASC foci/cell. Pie charts display the frequency of cells positive for biotin-YVAD-CMK staining per group. (d) Diameter of ASC foci in macrophages positive (filled green circles) and negative (unfilled circles) for biotin-YVAD-CMK staining from healthy control and SCAN4 patients II.3 and III.2. Mean foci diameter is displayed as white bars. Scale bars, 20 μm. An asterisk indicates that the patient cell distribution is significantly different (P = 0.0012) from the healthy control cell distribution by χ2 testing. Significance by unpaired Student’s t test is indicated (for d).
Supplementary Figure 6 ASC foci in S. typhimurium strain SL1344–infected SCAN4 macrophages do not colocalize with aggregates of caspase-1.
Uninfected macrophages from healthy controls (top row) do not form ASC foci (arrows), whereas S. typhimurium strain SL1344–infected healthy control macrophages (second row) stain diffusely with biotin-YVAD-CMK and form aggregates of caspase-1 (arrows) and ASC foci (arrows) that colocalize in merged images (arrows). SCAN4 macrophages diffusely staining negative (third row) or positive (fourth row) for biotin-YVAD-CMK form multiple ASC foci that do not colocalize with aggregates of caspase-1. DAPI staining is shown where labeled and also in the merged images. All images were taken at 60× magnification, except those for uninfected cells (top row), which were taken at 100× magnification. Scale bars are sized as indicated.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1–3. (PDF 2968 kb)
Rights and permissions
About this article
Cite this article
Romberg, N., Al Moussawi, K., Nelson-Williams, C. et al. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat Genet 46, 1135–1139 (2014). https://doi.org/10.1038/ng.3066
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3066
This article is cited by
-
The 4th NextGen therapies of SJIA and MAS, part 4: it is time for IL-18 based trials in systemic juvenile idiopathic arthritis?
Pediatric Rheumatology (2024)
-
Revealing novel pyroptosis-related therapeutic targets for sepsis based on machine learning
BMC Medical Genomics (2023)
-
Activation of distinct inflammatory pathways in subgroups of LR-MDS
Leukemia (2023)
-
Dietary Intake and Systemic Inflammation: Can We Use Food as Medicine?
Current Nutrition Reports (2023)
-
Circ_0000181 regulates miR-667-5p/NLRC4 axis to promote pyroptosis progression in diabetic nephropathy
Scientific Reports (2022)