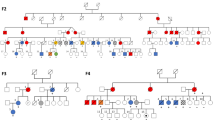
Abstract
We report germline missense mutations in ETV6 segregating with the dominant transmission of thrombocytopenia and hematologic malignancy in three unrelated kindreds, defining a new hereditary syndrome featuring thrombocytopenia with susceptibility to diverse hematologic neoplasms. Two variants, p.Arg369Gln and p.Arg399Cys, reside in the highly conserved ETS DNA-binding domain. The third variant, p.Pro214Leu, lies within the internal linker domain, which regulates DNA binding. These three amino acid sites correspond to hotspots for recurrent somatic mutation in malignancies. Functional studies show that the mutations abrogate DNA binding, alter subcellular localization, decrease transcriptional repression in a dominant-negative fashion and impair hematopoiesis. These familial genetic studies identify a central role for ETV6 in hematopoiesis and malignant transformation. The identification of germline predisposition to cytopenias and cancer informs the diagnosis and medical management of at-risk individuals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Sequence Read Archive
Referenced accessions
NCBI Reference Sequence
Protein Data Bank
References
Song, W.-J. et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23, 166–175 (1999).
Smith, M.L., Cavenagh, J.D., Lister, T.A. & Fitzgibbon, J. Mutation of CEBPA in familial acute myeloid leukemia. N. Engl. J. Med. 351, 2403–2407 (2004).
Hahn, C.N. et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 43, 1012–1017 (2011).
Kazenwadel, J. et al. Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood 119, 1283–1291 (2012).
Pippucci, T. et al. Mutations in the 5′ UTR of ANKRD26, the ankirin repeat domain 26 gene, cause an autosomal-dominant form of inherited thrombocytopenia, THC2. Am. J. Hum. Genet. 88, 115–120 (2011).
Noris, P. et al. Mutations in ANKRD26 are responsible for a frequent form of inherited thrombocytopenia: analysis of 78 patients from 21 families. Blood 117, 6673–6680 (2011).
Kirwan, M. et al. Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. Am. J. Hum. Genet. 90, 888–892 (2012).
Shah, S. et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat. Genet. 45, 1226–1231 (2013).
Auer, F. et al. Inherited susceptibility to pre B-ALL caused by germline transmission of PAX5 c.547G>A. Leukemia 28, 1136–1138 (2014).
Holmfeldt, L. et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 45, 242–252 (2013).
Powell, B.C. et al. Identification of TP53 as an acute lymphocytic leukemia susceptibility gene through exome sequencing. Pediatr. Blood Cancer 60, E1–E3 (2013).
Zhang, M.Y. et al. Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity. Haematologica 10.3324/haematol.2014.113456 (19 September 2014).
De, S. et al. Steric mechanism of auto-inhibitory regulation of specific and non-specific DNA binding by the ETS transcriptional repressor ETV6. J. Mol. Biol. 426, 1390–1406 (2014).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014).
Green, S.M., Coyne, H.J. III, McIntosh, L.P. & Graves, B.J. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J. Biol. Chem. 285, 18496–18504 (2010).
Coyne, H.J. et al. Autoinhibition of ETV6 (TEL) DNA binding: appended helices sterically block the ETS domain. J. Mol. Biol. 421, 67–84 (2012).
Chakrabarti, S.R. & Nucifora, G. The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochem. Biophys. Res. Commun. 264, 871–877 (1999).
Park, H., Seo, Y., Kim, J.I., Kim, W. & Choe, S.Y. Identification of the nuclear localization motif in the ETV6 (TEL) protein. Cancer Genet. Cytogenet. 167, 117–121 (2006).
Fenrick, R. et al. Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol. Cell. Biol. 19, 6566–6574 (1999).
Fenrick, R. et al. TEL, a putative tumor suppressor, modulates cell growth and cell morphology of Ras-transformed cells while repressing the transcription of stromelysin-1. Mol. Cell. Biol. 20, 5828–5839 (2000).
Lopez, R.G. et al. TEL is a sequence-specific transcriptional repressor. J. Biol. Chem. 274, 30132–30138 (1999).
Kwiatkowski, B.A. et al. The ets family member Tel binds to the Fli-1 oncoprotein and inhibits its transcriptional activity. J. Biol. Chem. 273, 17525–17530 (1998).
Kim, C.A. et al. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 20, 4173–4182 (2001).
Wang, L.C. et al. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 12, 2392–2402 (1998).
Hock, H. et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 18, 2336–2341 (2004).
Pritchard, C.C. et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J. Mol. Diagn. 16, 56–67 (2014).
Marquez, R. et al. A new family with a germline ANKRD26 mutation and predisposition to myeloid malignancies. Leuk. Lymphoma 10.3109/10428194.2014.903476 (22 April 2014).
Novershtern, N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309 (2011).
Bluteau, D. et al. Thrombocytopenia-associated mutations in the ANKRD26 regulatory region induce MAPK hyperactivation. J. Clin. Invest. 124, 580–591 (2014).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 (2012).
Welch, J.S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012).
Walter, M.J. et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia 27, 1275–1282 (2013).
Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012).
Bejar, R. et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 364, 2496–2506 (2011).
Xu, L. et al. Genomic landscape of CD34+ hematopoietic cells in myelodysplastic syndrome and gene mutation profiles as prognostic markers. Proc. Natl. Acad. Sci. USA 111, 8589–8594 (2014).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Dolnik, A. et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood 120, e83–e92 (2012).
Padron, E. et al. ETV6 and signaling gene mutations are associated with secondary transformation of myelodysplastic syndromes to chronic myelomonocytic leukemia. Blood 123, 3675–3677 (2014).
Griesinger, F., Janke, A., Podleschny, M. & Bohlander, S.K. Identification of an ETV6-ABL2 fusion transcript in combination with an ETV6 point mutation in a T-cell acute lymphoblastic leukaemia cell line. Br. J. Haematol. 119, 454–458 (2002).
Zhang, J. et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood 118, 3080–3087 (2011).
Wang, Q. et al. ETV6 mutation in a cohort of 970 patients with hematologic malignancies. Haematologica 99, e176–e178 (2014).
Lohr, J.G. et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25, 91–101 (2014).
Hodis, E. et al. A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012).
Walsh, T. et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of non-syndromic hearing loss DFNB82. Am. J. Hum. Genet. 87, 90–94 (2010).
Gulsuner, S. et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529 (2013).
Walsh, T. et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. USA 108, 18032–18037 (2011).
Mostoslavsky, G., Fabian, A.J., Rooney, S., Alt, F.W. & Mulligan, R.C. Complete correction of murine Artemis immunodeficiency by lentiviral vector–mediated gene transfer. Proc. Natl. Acad. Sci. USA 103, 16406–16411 (2006).
Delaney, C., Varnum-Finney, B., Aoyama, K., Brashem-Stein, C. & Bernstein, I.D. Dose-dependent effects of the Notch ligand Delta1 on ex vivo differentiation and in vivo marrow repopulating ability of cord blood cells. Blood 106, 2693–2699 (2005).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Anders, S., Pyl, P.T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics doi:10.1093/bioinformatics/btu638 (25 September 2014).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Reiner, A., Yekutieli, D. & Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 (2003).
Yeung, K.Y., Haynor, D.R. & Ruzzo, W.L. Validating clustering for gene expression data. Bioinformatics 17, 309–318 (2001).
Saeed, A.I. et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378 (2003).
Young, M.D., Wakefield, M.J., Smyth, G.K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Burwick, N., Coats, S.A., Nakamura, T. & Shimamura, A. Impaired ribosomal subunit association in Shwachman-Diamond syndrome. Blood 120, 5143–5152 (2012).
Acknowledgements
We thank all patients and their families for participation in this research study. We thank M. Chin (University of Washington), B. Turok-Storb (Fred Hutchinson Cancer Research Center) and S. Tapscott (Fred Hutchinson Cancer Research Center) for luciferase plasmids and reagents. We thank H. Hock, B. Stoddard, S. Meshinchi, G. Smith, A. Kumar, C. Toledo, S. Yu, A. Fong and K. MacQuarrie for helpful discussions. We thank S. Castro for clinical sample processing. This work was supported by US National Institutes of Health grants R24DK093425 and R24DK099808-01 to A.S., M.-C.K. and J.L.A.; by the Ghiglione Aplastic Anemia Fund and Julian's Dinosaur Guild from Seattle Children's Hospital to A.S.; by Medical Scientist Training Program Training grant T32GM007266 and Genetic Approaches to Aging Training grant T32AG000057 to M.Y.Z.; and by grants from the US National Institutes of Health (K12CA139160) and the Cancer Research Foundation to J.E.C. M.-C.K. is an American Cancer Society professor.
Author information
Authors and Affiliations
Contributions
M.Y.Z., J.E.C., S.B.K., T.W., J.L.A., M.-C.K., L.A.G. and A.S. conceived and designed the experiments. M.Y.Z., S.B.K., T.W., C.C.P., M.S.-B., C.J.M. and S.A.C. performed the experiments. M.Y.Z., S.B.K., T.W., M.K.L., K.R.L., S.G., C.C.P., J.J.D., R.S.B., R.C.L., M.-C.K. and A.S. analyzed the data. J.E.C., S.B.K., M.F., B.G., B.S.S., B.N., R.M., I.H., D.A.W., M.S.H., L.A.G. and A.S. identified study subjects, performed clinical phenotyping and contributed biological samples. M.Y.Z., J.E.C., T.W., J.J.D., L.A.G., M.-C.K. and A.S. wrote the manuscript. A.S. and M.-C.K. jointly supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Bone marrow morphology in III-2 of family A.
(a) Pre-transplant peripheral blood smears from patient III-2 illustrating hypogranulated neutrophils (left) and hypolobated neutrophils (pseudo Pelger-Huët cell) (right). (b) Platelets from a Wright-Giemsa stained peripheral blood smear. (c) Upper left, Wright-Giemsa stained particle preparation depicting small, hypolobated megakaryocytes (triangles). Lower left panel: Bone marrow biopsy depicting hypolobated micro-megakaryocytes. Right, megakaryocyte dysplasia.
Supplementary Figure 2 Sanger sequencing of the ETV6 c.1195C>T mutation in family A.
(a) Electropherograms from Sanger sequencing of ETV6 c.1195C>T in peripheral blood genomic DNA from family A. (b) Sanger sequencing of ETV6 c.1195C>T in genomic DNA and cDNA derived from an RAEB-1 MDS bone marrow sample of family A III-2 shows no loss of heterozygosity.
Supplementary Figure 3 The ETV6 p.Arg369Gln mutation disrupts internal hydrogen bonding.
(a) Hydrogen bonding (dotted lines) between the guanidinium nitrogen of Arg369 (orange) in β sheet 2 with the backbone carbonyl oxygen of Arg414 (magenta) in the wing of the ETS domain. Protein structure of the murine Etv6 ETS domain (PDB ID: 4MHG) is shown. The ETS domains of mouse and human ETV6 have 100% amino acid sequence identity. (b) Molecular modeling of the Arg369Gln (orange) variant using SWISS-MODEL predicts loss of this hydrogen bonding interaction.
Supplementary Figure 4 Cell fractionation shows decreased nuclear localization of mutant ETV6.
HeLa cells were transfected with empty vector, cDNA for wild-type ETV6 or ETV6 cDNA encoding p.Arg399Cys. Lysates of nuclear versus cell fractions as well as whole-cell lysates were analyzed by western blot for ETV6, GAPDH (cytoplasmic marker) and NPM1 (nuclear marker).
Supplementary Figure 5 Clonal evolution of MDS in the context of germline ETV6 mutation.
Analysis of germline and serial bone marrow samples in family A III-2. Electropherograms from Sanger sequencing of BCOR, RUNX1, and KRAS mutations in genomic DNA derived from (a) marrow fibroblasts, (b) bone marrow mononuclear cells at age 17 when the patient had refractory cytopenias with multilineage dysplasia (RCMD) and (c) bone marrow mononuclear cells at age 21 when the patient’s disease progressed to refractory anemia with excess blasts type 1 (RAEB-1).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–3 and 8–10, and Supplementary Note. (PDF 2630 kb)
Supplementary Table 4
Genes downregulated with mutant ETV6 compared to WT ETV6. (XLSX 70 kb)
Supplementary Table 5
Genes upregulated with mutant ETV6 compared to WT ETV6. (XLSX 76 kb)
Supplementary Table 6
GO-seq categories for genes downregulated with mutant ETV6 compared to WT ETV6. (XLSX 77 kb)
Supplementary Table 7
GO-seq categories for genes upregulated with mutant ETV6 compared to WT ETV6. (XLSX 46 kb)
Rights and permissions
About this article
Cite this article
Zhang, M., Churpek, J., Keel, S. et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet 47, 180–185 (2015). https://doi.org/10.1038/ng.3177
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3177