Abstract
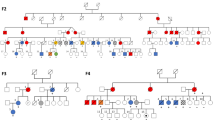
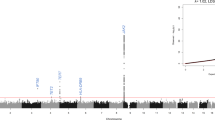
Polycythemia vera, essential thrombocythemia and primary myelofibrosis are myeloproliferative neoplasms (MPN) characterized by multilineage clonal hematopoiesis1,2,3,4,5. Given that the identical somatic activating mutation in the JAK2 tyrosine kinase gene (JAK2V617F) is observed in most individuals with polycythemia vera, essential thrombocythemia and primary myelofibrosis6,7,8,9,10, there likely are additional genetic events that contribute to the pathogenesis of these phenotypically distinct disorders. Moreover, family members of individuals with MPN are at higher risk for the development of MPN, consistent with the existence of MPN predisposition loci11. We hypothesized that germline variation contributes to MPN predisposition and phenotypic pleiotropy. Genome-wide analysis identified an allele in the JAK2 locus (rs10974944) that predisposes to the development of JAK2V617F-positive MPN, as well as three previously unknown MPN modifier loci. We found that JAK2V617F is preferentially acquired in cis with the predisposition allele. These data suggest that germline variation is an important contributor to MPN phenotype and predisposition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Adamson, J.W., Fialkow, P.J., Murphy, S., Prchal, J.F. & Steinmann, L. Polycythemia vera: stem-cell and probable clonal origin of the disease. N. Engl. J. Med. 295, 913–916 (1976).
Gilliland, D.G., Blanchard, K.L., Levy, J., Perrin, S. & Bunn, H.F. Clonality in myeloproliferative disorders: analysis by means of the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 88, 6848–6852 (1991).
El Kassar, N., Hetet, G., Li, Y., Briere, J. & Grandchamp, B. Clonal analysis of haemopoietic cells in essential thrombocythaemia. Br. J. Haem. 90, 131–137 (1995).
Tsukamoto, N. et al. Clonality in chronic myeloproliferative disorders defined by X-chromosome linked probes: demonstration of heterogeneity in lineage involvement. Br. J. Haematol. 86, 253–258 (1994).
Jamieson, C.H. et al. The JAK2 V617F mutation occurs in hematopoietic stem cells in polycythemia vera and predisposes toward erythroid differentiation. Proc. Natl. Acad. Sci. USA 103, 6224–6229 (2006).
James, C. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005).
Baxter, E.J. et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061 (2005).
Kralovics, R. et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790 (2005).
Zhao, R. et al. Identification of an acquired JAK2 mutation in polycythemia vera. J. Biol. Chem. 280, 22788–22792 (2005).
Levine, R.L. et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7, 387–397 (2005).
Landgren, O. et al. Increased risks of polycythemia vera, essential thrombocythemia, and myelofibrosis among 24577 first-degree relatives of 11039 patients with myeloproliferative neoplasms in Sweden. Blood 112, 2199–2204 (2008).
Pardanani, A., Fridley, B.L., Lasho, T.L., Gilliland, D.G. & Tefferi, A. Host genetic variation contributes to phenotypic diversity in myeloproliferative disorders. Blood 111, 2785–2789 (2008).
Cario, H., Goerttler, P.S., Steimle, C., Levine, R.L. & Pahl, H.L. The JAK2V617F mutation is acquired secondary to the predisposing alteration in familial polycythaemia vera. Br. J. Haematol. 130, 800–801 (2005).
Bellanne-Chantelot, C. et al. Genetic and clinical implications of the Val617Phe JAK2 mutation in 72 families with myeloproliferative disorders. Blood 108, 346–352 (2006).
Pietra, D. et al. Somatic mutations of JAK2 exon 12 in patients with JAK2 (V617F)-negative myeloproliferative disorders. Blood 111, 1686–1689 (2008).
Scott, L.M., Scott, M.A., Campbell, P.J. & Green, A.R. Progenitors homozygous for the V617F mutation occur in most patients with polycythemia vera, but not essential thrombocythemia. Blood 108, 2435–2437 (2006).
Scott, L.M. et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N. Engl. J. Med. 356, 459–468 (2007).
Di Bernardo, M.C. et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 40, 1204–1210 (2008).
Hemminki, K., Forsti, A. & Bermejo, J.L. The 'common disease-common variant' hypothesis and familial risks. PLoS ONE 3, e2504 (2008).
Hemminki, K., Forsti, A. & Lorenzo Bermejo, J. New cancer susceptibility loci: population and familial risks. Int. J. Cancer 123, 1726–1729 (2008).
Laken, S.J. et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat. Genet. 17, 79–83 (1997).
Bercovich, D. et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet 372, 1484–1492 (2008).
Mercher, T. et al. JAK2T875N is a novel activating mutation that results in myeloproliferative disease with features of megakaryoblastic leukemia in a murine bone marrow transplantation model. Blood 108, 2770–2779 (2006).
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Levine, R.L. et al. X-inactivation-based clonality analysis and quantitative JAK2V617F assessment reveal a strong association between clonality and JAK2V617F in PV but not ET/MMM, and identifies a subset of JAK2V617F-negative ET and MMM patients with clonal hematopoiesis. Blood 107, 4139–4141 (2006).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Acknowledgements
We would like to acknowledge the subjects who have contributed to our understanding of these disorders. We thank S. Thomas, I. Dolgalev and T. Landers for assistance with high-throughput resequencing, A. Viale for assistance with JAK2 expression analysis, and T. Kirchhoff for advice and suggestions. This study makes use of data generated by the Wellcome Trust Case-Control Consortium; a full list of the investigators who contributed to the generation of the data are available from http://www.wtccc.org.uk and funding was provided by the Wellcome Trust under award 076113. This work was supported by grants from the National Institutes of Health, the Starr Cancer Consortium, the Myeloproliferative Disorders Foundation, the Howard Hughes Medical Institute, the Doris Duke Charitable Foundation and the Kristen Amico Sesselman Leukemia Research Fund. O.K. is supported by a grant from the Academy of Finland. D.G.G. is an Investigator of the Howard Hughes Medical Institute and is a Doris Duke Charitable Foundation Distinguished Clinical Scientist. Work in the laboratory of R.J.K. is supported by Memorial Sloan Kettering Cancer Center through US National Institutes of Health grant P30 CA008748. R.L.L. is an Early Career Award recipient of the Howard Hughes Medical Institute and a Clinical Scientist Development Award recipient of the Doris Duke Charitable Foundation and is the Geoffrey Beene Junior Chair at Memorial Sloan Kettering Cancer Center.
Author information
Authors and Affiliations
Contributions
The study was designed by O.K., S. Mukherjee, R.J.K. and R.L.L. with advice from K.O. SNP arrays were performed and analyzed by A.B., B.L.E. and R.L.L, and analysis of SNP array data for modifier and predisposition loci was performed by S. Mukherjee and R.J.K. Genotyping, sequence analysis and real-time PCR assays were performed by O.K., A.M.S., S. Marubayashi, A.H. and R.L.L. Principal component analysis was done by S. Mukherjee and R.J.K. Identification of subjects, sample collection and phenotypic assessment were done by M.W., A.M., G.G.-M., H.K., R.M.S, D.G.G. and R.L.L. The paper was written by O.K., S. Mukherjee, K.O., D.G.G., R.J.K. and R.L.L. All authors discussed the results and commented on the manuscript.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Tables 1–3 (PDF 336 kb)
Rights and permissions
About this article
Cite this article
Kilpivaara, O., Mukherjee, S., Schram, A. et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2V617F-positive myeloproliferative neoplasms. Nat Genet 41, 455–459 (2009). https://doi.org/10.1038/ng.342
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.342
This article is cited by
-
PASTRY: achieving balanced power for detecting risk and protective minor alleles in meta-analysis of association studies with overlapping subjects
BMC Bioinformatics (2024)
-
The aetiology and burden of myeloproliferative neoplasms in the United Kingdom: the MyelOproliferative neoplasmS: an In-depth case-control (MOSAICC) study protocol
BMC Cancer (2023)
-
Molecular Pathogenesis of Myeloproliferative Neoplasms
Current Hematologic Malignancy Reports (2022)
-
Germline-somatic JAK2 interactions are associated with clonal expansion in myelofibrosis
Nature Communications (2022)
-
Advances in germline predisposition to acute leukaemias and myeloid neoplasms
Nature Reviews Cancer (2021)