Abstract
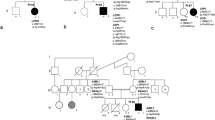
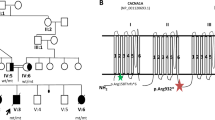
We report germline loss-of-function mutations in SPRED1 in a newly identified autosomal dominant human disorder. SPRED1 is a member of the SPROUTY/SPRED family1 of proteins that act as negative regulators of RAS->RAF interaction and mitogen-activated protein kinase (MAPK) signaling2. The clinical features of the reported disorder resemble those of neurofibromatosis type 1 and consist of multiple café-au-lait spots, axillary freckling and macrocephaly. Melanocytes from a café-au-lait spot showed, in addition to the germline SPRED1 mutation, an acquired somatic mutation in the wild-type SPRED1 allele, indicating that complete SPRED1 inactivation is needed to generate a café-au-lait spot in this syndrome. This disorder is yet another member of the recently characterized group of phenotypically overlapping syndromes caused by mutations in the genes encoding key components of the RAS-MAPK pathway3,4. To our knowledge, this is the first report of mutations in the SPRY (SPROUTY)/SPRED family of genes in human disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bundschu, K., Walter, U. & Schuh, K. The VASP-Spred-Sprouty domain puzzle. J. Biol. Chem. 281, 36477–36481 (2006).
Wakioka, T. et al. Spred is a Sprouty-related suppressor of Ras signalling. Nature 412, 647–651 (2001).
Shannon, K. & Bollag, G. Sending out an SOS. Nat. Genet. 39, 8–9 (2007).
Bentires-Alj, M., Kontaridis, M.I. & Neel, B.G. Stops along the RAS pathway in human genetic disease. Nat. Med. 12, 283–285 (2006).
Legius, E., Marchuk, D.A., Collins, F.S. & Glover, T.W. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat. Genet. 3, 122–126 (1993).
Cichowski, K. & Jacks, T. NF1 tumor suppressor gene function: narrowing the GAP. Cell 104, 593–604 (2001).
Tartaglia, M. et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29, 465–468 (2001).
Schubbert, S. et al. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 38, 331–336 (2006).
Roberts, A.E. et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 39, 70–74 (2007).
Tartaglia, M. et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 39, 75–79 (2007).
Digilio, M.C. et al. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am. J. Hum. Genet. 71, 389–394 (2002).
Legius, E. et al. PTPN11 mutations in LEOPARD syndrome. J. Med. Genet. 39, 571–574 (2002).
Aoki, Y. et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 37, 1038–1040 (2005).
Niihori, T. et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 38, 294–296 (2006).
Rodriguez-Viciana, P. et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311, 1287–1290 (2006).
Messiaen, L.M. et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum. Mutat. 15, 541–555 (2000).
De Schepper, S. et al. Cafe-au-lait spots in neurofibromatosis type 1 and in healthy control individuals: hyperpigmentation of a different kind? Arch. Dermatol. Res. 297, 439–449 (2006).
Gille, H. et al. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 14, 951–962 (1995).
King, J.A.J. et al. Distinct requirements for the Sprouty domain for functional activity of Spred proteins. Biochem. J. 388, 445–454 (2005).
Miyoshi, K. et al. The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene 23, 5567–5576 (2004).
Engelhardt, C.M. et al. Expression and subcellular localization of Spred proteins in mouse and human tissues. Histochem. Cell Biol. 122, 527–538 (2004).
Inoue, H. et al. Spred-1 negatively regulates allergen-induced airway eosinophilia and hyperresponsiveness. J. Exp. Med. 201, 73–82 (2005).
Stumpf, D. et al. Neurofibromatosis conference statement. National Institutes of Health Consensus Development Conference. Arch. Neurol. 45, 575–578 (1988).
Maertens, O. et al. Molecular dissection of isolated disease features in mosaic neurofibromatosis type 1. Am. J. Hum. Genet. 81, 243–251 (2007).
Taniguchi, K. et al. Spreds are essential for embryonic lymphangiogenesis by regulating vascular endothelial growth factor receptor 3 signaling. Mol. Cell. Biol. 27, 4541–4550 (2007).
Sasaki, A. et al. Identification of a dominant negative mutant of Sprouty that potentiates fibroblast growth factor- but not epidermal growth factor-induced ERK activation. J. Biol. Chem. 276, 36804–36808 (2001).
Acknowledgements
The authors thank T. de Ravel for critically reading the manuscript, M. De Mil for technical assistance in melanocyte cell culture and F. Okamoto for technical assistance in cell culture and plasmid preparation. H.B. is supported by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen). M.C. was supported by the Marie Curie European Community fellowship (contract HPMT-CT2001-00273). E.D. is a predoctoral researcher (Aspirant of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen), J.C. is a postdoctoral researcher and E.L. is a part-time clinical researcher of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (FWO). This work is also supported by research grants from the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (G.0096.02, E.L.) and (G.0507.04, P.M.); by the Interuniversity Attraction Poles (IAP) granted by the Federal Office for Scientific, Technical and Cultural Affairs, Belgium (2002–2006; P5/25) (P.M. and E.L.); by a Concerted Action Grant from the Catholic University Leuven; by the Federation des Maladies Genetiques Orphelines (G.T.) and by grants-in-aid from the Ministry of Education, Science, Technology, Sports, and Culture of Japan (A.Y.). This work was made possible by the Centre of Excellence SymBioSys (Research Council, Catholic University Leuven EF/05/007) (E.L.).
Author information
Authors and Affiliations
Contributions
The study was coordinated by H.B., G.T., P.M., A.Y. and E.L.; patient phenotyping was performed by J.-P.F. and E.L. and clinical data collected by E.D.; linkage analysis was done by M.S. and G.T.; NF1 mutation analysis was conducted by L.M. and SPRED1 mutation analysis by H.B., M.C. and E.D.; skin biopsies were carried out by S.D.S. and melanocytes cultured by H.B. and S.D.S.; biochemical analysis was performed by H.B., M.C., E.D., K.T., R.S. and J.C.; mouse characterization was performed by R.K.; the manuscript was written by H.B., M.C., E.D., L.M., S.D.S., A.Y., J.-P.F., J.C., P.M., G.T. and E.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Table 1, Supplementary Figures 1–7 (PDF 635 kb)
Rights and permissions
About this article
Cite this article
Brems, H., Chmara, M., Sahbatou, M. et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1–like phenotype. Nat Genet 39, 1120–1126 (2007). https://doi.org/10.1038/ng2113
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng2113
This article is cited by
-
Novel cellular systems unveil mucosal melanoma initiating cells and a role for PI3K/Akt/mTOR pathway in mucosal melanoma fitness
Journal of Translational Medicine (2024)
-
SPOCK2 and SPRED1 function downstream of EZH2 to impede the malignant progression of lung adenocarcinoma in vitro and in vivo
Human Cell (2023)
-
Neurofibromatosis type1, type 2, tuberous sclerosis and Von Hippel-Lindau disease
Child's Nervous System (2023)
-
Spred1 deficit promotes treatment resistance and transformation of chronic phase CML
Leukemia (2022)
-
Molecular and clinical profile of patients referred as Noonan or Noonan-like syndrome in Greece: a cohort of 86 patients
European Journal of Pediatrics (2022)