Abstract
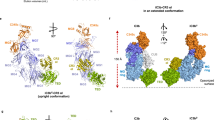
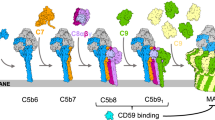
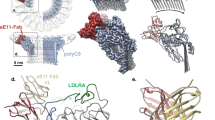
Activation of the complement system generates potent chemoattractants and leads to the opsonization of cells for immune clearance. Short-lived protease complexes cleave complement component C3 into anaphylatoxin C3a and opsonin C3b. Here we report the crystal structure of the C3 convertase formed by C3b and the protease fragment Bb, which was stabilized by the bacterial immune-evasion protein SCIN. The data suggest that the proteolytic specificity and activity depend on the formation of dimers of C3 with C3b of the convertase. SCIN blocked the formation of a productive enzyme-substrate complex. Irreversible dissociation of the complex of C3b and Bb is crucial to complement regulation and was determined by slow binding kinetics of the Mg2+-adhesion site in Bb. Understanding the mechanistic basis of the central complement-activation step and microbial immune evasion strategies targeting this step will aid in the development of complement therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Carroll, M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 5, 981–986 (2004).
Mollnes, T.E., Song, W.C. & Lambris, J.D. Complement in inflammatory tissue damage and disease. Trends Immunol. 23, 61–64 (2002).
Duncan, R.C., Wijeyewickrema, L.C. & Pike, R.N. The initiating proteases of the complement system: controlling the cleavage. Biochimie 90, 387–395 (2008).
Law, S.K. & Dodds, A.W. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 6, 263–274 (1997).
Gros, P., Milder, F.J. & Janssen, B.J. Complement driven by conformational changes. Nat. Rev. Immunol. 8, 48–58 (2008).
Kerr, M.A. The human complement system: assembly of the classical pathway C3 convertase. Biochem. J. 189, 173–181 (1980).
Rawal, N. & Pangburn, M.K. C5 convertase of the alternative pathway of complement. Kinetic analysis of the free and surface-bound forms of the enzyme. J. Biol. Chem. 273, 16828–16835 (1998).
Kinoshita, T. et al. C5 convertase of the alternative complement pathway: covalent linkage between two C3b molecules within the trimolecular complex enzyme. J. Immunol. 141, 3895–3901 (1988).
Rawal, N. & Pangburn, M.K. Structure/function of C5 convertases of complement. Int. Immunopharmacol. 1, 415–422 (2001).
Kirkitadze, M.D. & Barlow, P.N. Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol. Rev. 180, 146–161 (2001).
Pangburn, M.K. & Muller-Eberhard, H.J. The C3 convertase of the alternative pathway of human complement. Enzymic properties of the bimolecular proteinase. Biochem. J. 235, 723–730 (1986).
Lambris, J.D., Ricklin, D. & Geisbrecht, B.V. Complement evasion by human pathogens. Nat. Rev. Microbiol. 6, 132–142 (2008).
Rooijakkers, S.H. et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6, 920–927 (2005).
Rooijakkers, S.H. et al. Staphylococcal complement inhibitor: structure and active sites. J. Immunol. 179, 2989–2998 (2007).
Janssen, B.J., Christodoulidou, A., McCarthy, A., Lambris, J.D. & Gros, P. Structure of C3b reveals conformational changes that underlie complement activity. Nature 444, 213–216 (2006).
Wiesmann, C. et al. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature 444, 217–220 (2006).
Torreira, E., Tortajada, A., Montes, T., de Cordoba, S.R. & Llorca, O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proc. Natl. Acad. Sci. USA 106, 882–887 (2009).
Muller-Eberhard, H.J. & Gotze, O. C3 proactivator convertase and its mode of action. J. Exp. Med. 135, 1003–1008 (1972).
Fishelson, Z., Pangburn, M.K. & Muller-Eberhard, H.J. C3 convertase of the alternative complement pathway. Demonstration of an active, stable C3 C3b,Bb (Ni) complex. J. Biol. Chem. 258, 7411–7415 (1983).
Horiuchi, T., Macon, K.J., Engler, J.A. & Volanakis, J.E. Site-directed mutagenesis of the region around Cys-241 of complement component C2. Evidence for a C4b binding site. J. Immunol. 147, 584–589 (1991).
Hourcade, D.E., Mitchell, L.M. & Oglesby, T.J. Mutations of the type A domain of complement factor B that promote high-affinity C3b-binding. J. Immunol. 162, 2906–2911 (1999).
Tuckwell, D.S., Xu, Y., Newham, P., Humphries, M.J. & Volanakis, J.E. Surface loops adjacent to the cation-binding site of the complement factor B von Willebrand factor type A module determine C3b binding specificity. Biochemistry 36, 6605–6613 (1997).
Hourcade, D.E., Mitchell, L., Kuttner-Kondo, L.A., Atkinson, J.P. & Medof, M.E. Decay-accelerating factor (DAF), complement receptor 1 (CR1), and factor H dissociate the complement AP C3 convertase (C3bBb) via sites on the type A domain of Bb. J. Biol. Chem. 277, 1107–1112 (2002).
Ponnuraj, K. et al. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol. Cell 14, 17–28 (2004).
Milder, F.J. et al. Factor B structure provides insights into activation of the central protease of the complement system. Nat. Struct. Mol. Biol. 14, 224–228 (2007).
Milder, F.J. et al. Structure of complement component C2a: implications for convertase formation and substrate binding. Structure 14, 1587–1597 (2006).
Luo, B.H., Carman, C.V. & Springer, T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 (2007).
Krishnan, V., Xu, Y., Macon, K., Volanakis, J.E. & Narayana, S.V. The crystal structure of C2a, the catalytic fragment of classical pathway C3 and C5 convertase of human complement. J. Mol. Biol. 367, 224–233 (2007).
Kam, C.M. et al. Human complement proteins D, C2, and B. Active site mapping with peptide thioester substrates. J. Biol. Chem. 262, 3444–3451 (1987).
Janssen, B.J., Halff, E.F., Lambris, J.D. & Gros, P. Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J. Biol. Chem. 282, 29241–29247 (2007).
Katschke, K.J. Jr. et al. Structural and functional analysis of a C3b-specific antibody that selectively inhibits the alternative pathway of complement. J. Biol. Chem. 284, 10473–10479 (2009).
Janssen, B.J. et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 437, 505–511 (2005).
Fredslund, F. et al. Structure of and influence of a tick complement inhibitor on human complement component 5. Nat. Immunol. 9, 753–760 (2008).
Rawal, N. & Pangburn, M. Formation of high-affinity C5 convertases of the alternative pathway of complement. J. Immunol. 166, 2635–2642 (2001).
Hourcade, D.E., Mitchell, L.M. & Medof, M.E. Decay acceleration of the complement alternative pathway C3 convertase. Immunopharmacology 42, 167–173 (1999).
Bhattacharya, A.A., Lupher, M.L., Jr., Staunton, D.E. & Liddington, R.C. Crystal structure of the A domain from complement factor B reveals an integrin-like open conformation. Structure 12, 371–378 (2004).
Harris, C.L., Abbott, R.J., Smith, R.A., Morgan, B.P. & Lea, S.M. Molecular dissection of interactions between components of the alternative pathway of complement and decay accelerating factor (CD55). J. Biol. Chem. 280, 2569–2578 (2005).
Pryzdial, E.L. & Isenman, D.E. Alternative complement pathway activation fragment Ba binds to C3b. Evidence that formation of the factor B-C3b complex involves two discrete points of contact. J. Biol. Chem. 262, 1519–1525 (1987).
Lambris, J.D., Dobson, N.J. & Ross, G.D. Release of endogenous C3b inactivator from lymphocytes in response to triggering membrane receptors for β1H globulin. J. Exp. Med. 152, 1625–1644 (1980).
Reeves, P.J., Callewaert, N., Contreras, R. & Khorana, H.G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA 99, 13419–13424 (2002).
de Haas, C.J. et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 199, 687–695 (2004).
Demeler, B. & van Holde, K.E. Sedimentation velocity analysis of highly heterogeneous systems. Anal. Biochem. 335, 279–288 (2004).
Demeler, B. in Analytical Ultracentrifugation: Techniques and Methods (eds. Scott, D.J., Harding S.E. & Rowe, A.J.) 210–230 (Royal Society of Chemistry, Cambridge, 2005).
Brookes, E. & Demeler, B. Parallel computational techniques for the analysis of sedimentation velocity experiments in UltraScan. Colloid Polym. Sci. 286, 139–148 (2008).
Evans, P. Scaling and assessment of data quality. Acta Crystallographica Section D 62, 72–82 (2006).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Myszka, D.G. & Morton, T.A. CLAMP: a biosensor kinetic data analysis program. Trends Biochem. Sci. 23, 149–150 (1998).
Acknowledgements
We thank R. Romijn for help with mammalian protein expression; M. Otten and M. Daha for doing hemolytic assays; P. Lenting for help with Biacore analyses; the European Synchrotron Radiation Facility for synchrotron radiation facilities; and beamline scientists of the European Synchrotron Radiation Facility and the European Molecular Biology Laboratory for assistance. Supported by the Councils for Medical Sciences and Chemical Sciences of the Netherlands Organization for Scientific Research (S.H.M.R., J.A.G.v.S. and P.G.) and the US National Institutes of Health (J.D.L. and P.G.).
Author information
Authors and Affiliations
Contributions
J.W. expressed and purified FB and FD; M.R. and S.H.M.R. expressed and purified SCIN and chimeras; M.R. purified C3b; R.v.D., M.R. and S.H.M.R. generated and analyzed complexes and did functional assays; K.L.P. did and analyzed the analytical ultracentrifugation experiments; J.W. crystallized the complex and determined and analyzed the structure; B.J.C.J. helped with structure determination and analysis; A.T. expressed and purified Ba; D.R. did the C3b-FB and C3b-Ba binding studies; S.H.M.R., B.J.C.J., J.W., J.D.L., J.A.G.v.S. and P.G. conceived the experiments; and J.W., S.H.M.R., D.R., J.A.G.v.S. and P.G. wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 and Supplementary Table 1 (PDF 1183 kb)
Rights and permissions
About this article
Cite this article
Rooijakkers, S., Wu, J., Ruyken, M. et al. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol 10, 721–727 (2009). https://doi.org/10.1038/ni.1756
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1756
This article is cited by
-
Insight into mode-of-action and structural determinants of the compstatin family of clinical complement inhibitors
Nature Communications (2022)
-
Invariant surface glycoprotein 65 of Trypanosoma brucei is a complement C3 receptor
Nature Communications (2022)
-
Paracetamol modulates biofilm formation in Staphylococcus aureus clonal complex 8 strains
Scientific Reports (2021)
-
Complement Evasion Strategies of Human Pathogenic Bacteria
Indian Journal of Microbiology (2020)
-
How novel structures inform understanding of complement function
Seminars in Immunopathology (2018)