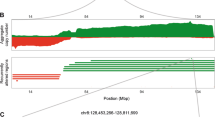
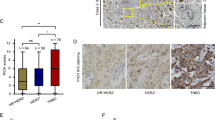
Abstract
Adjuvant chemotherapy for breast cancer after surgery has effectively lowered metastatic recurrence rates1. However, a considerable proportion of women suffer recurrent cancer at distant metastatic sites despite adjuvant treatment. Identification of the genes crucial for tumor response to specific chemotherapy drugs is a challenge but is necessary to improve outcomes2. By using integrated genomics, we identified a small number of overexpressed and amplified genes from chromosome 8q22 that were associated with early disease recurrence despite anthracycline-based adjuvant chemotherapy. We confirmed the association in an analysis of multiple independent cohorts. SiRNA-mediated knockdown of either of two of these genes, the antiapoptotic gene YWHAZ and a lysosomal gene LAPTM4B, sensitized tumor cells to anthracyclines, and overexpression of either of the genes induced anthracycline resistance. Overexpression of LAPTM4B resulted in sequestration of the anthracycline doxorubicin, delaying its appearance in the nucleus. Overexpression of these two genes was associated with poor tumor response to anthracycline treatment in a neoadjuvant chemotherapy trial in women with primary breast cancer. Our results suggest that 8q22 amplification and overexpression of LAPTM4B and YWHAZ contribute to de novo chemoresistance to anthracyclines and are permissive for metastatic recurrence. Overexpression of these two genes may predict anthracycline resistance and influence selection of chemotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365, 1687–1717 (2005).
Dowsett, M. et al. International Web-based consultation on priorities for translational breast cancer research. Breast Cancer Res. 9, R81 (2007).
Gottesman, M.M., Fojo, T. & Bates, S.E. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58 (2002).
Turton, N.J. et al. Gene expression and amplification in breast carcinoma cells with intrinsic and acquired doxorubicin resistance. Oncogene 20, 1300–1306 (2001).
Potti, A. et al. Genomic signatures to guide the use of chemotherapeutics. Nat. Med. 12, 1294–1300 (2006).
Györffy, B. et al. Prediction of doxorubicin sensitivity in breast tumors based on gene expression profiles of drug-resistant cell lines correlates with patient survival. Oncogene 24, 7542–7551 (2005).
Lee, J.K. et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc. Natl. Acad. Sci. USA 104, 13086–13091 (2007).
Liedtke, C. et al. Clinical evaluation of chemotherapy response predictors developed from breast cancer cell lines. Breast Cancer Res. Treat. published online, doi:10.1007/s10549-009-0445-7 (15 July 2009).
Ingvarsson, S. Molecular genetics of breast cancer progression. Semin. Cancer Biol. 9, 277–288 (1999).
Albertson, D.G. Profiling breast cancer by array CGH. Breast Cancer Res. Treat. 78, 289–298 (2003).
Albertson, D.G., Collins, C., McCormick, F. & Gray, J.W. Chromosome aberrations in solid tumors. Nat. Genet. 34, 369–376 (2003).
Chin, K. et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10, 529–541 (2006).
Tibshirani, R., Hastie, T., Narasimhan, B. & Chu, G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 99, 6567–6572 (2002).
Wang, Y. et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 (2005).
van 't Veer, L.J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
Hu, G. et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell 15, 9–20 (2009).
van de Vijver, M.J. et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002).
Ivshina, A.V. et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 66, 10292–10301 (2006).
Sotiriou, C. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98, 262–272 (2006).
Pawitan, Y. et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 7, R953–R964 (2005).
Niemantsverdriet, M., Wagner, K., Visser, M. & Backendorf, C. Cellular functions of 14–3-3ζ in apoptosis and cell adhesion emphasize its oncogenic character. Oncogene 27, 1315–1319 (2007).
Neal, C.L. et al. 14–3-3ζ overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 69, 3425–3432 (2009).
Zhao, J.J. et al. The oncogenic properties of mutant p110α and p110β phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc. Natl. Acad. Sci. USA 102, 18443–18448 (2005).
Zweig, M.H. & Campbell, G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin. Chem. 39, 561–577 (1993).
Farmer, P. et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat. Med. 15, 68–74 (2009).
Silver, D.P. et al. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J. Clin. Oncol. (in the press).
Chang, J.C. et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet 362, 362–369 (2003).
Hogue, D.L., Kerby, L. & Ling, V. A mammalian lysosomal membrane protein confers multidrug resistance upon expression in Saccharomyces cerevisiae. J. Biol. Chem. 274, 12877–12882 (1999).
Matros, E. et al. BRCA1 promoter methylation in sporadic breast tumors: relationship to gene expression profiles. Breast Cancer Res. Treat. 91, 179–186 (2005).
Lu, X. et al. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res. Treat. 108, 191–201 (2008).
Richardson, A.L. et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9, 121–132 (2006).
Wang, Z.C. et al. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res. 64, 64–71 (2004).
Desmedt, C. et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin. Cancer Res. 14, 5158–5165 (2008).
Harrell, F.E. Jr., Califf, R.M., Pryor, D.B., Lee, K.L. & Rosati, R.A. Evaluating the yield of medical tests. J. Am. Med. Assoc. 247, 2543–2546 (1982).
Pencina, M.J. & D'Agostino, R.B. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat. Med. 23, 2109–2123 (2004).
van Vliet, M.H. et al. Pooling breast cancer datasets has a synergetic effect on classification performance and improves signature stability. BMC Genomics 9, 375 (2008).
Acknowledgements
We thank C. Lee, A. Aggarwal, E. Fox, P. Hollasch, M. Berkeley, R. Gelman, W. Luo, X. Lu and members of the Richardson-Wang lab for their advice and assistance. We also thank J.E. Garber, A.C. Eklund, N. Juul and R.-L. Zhou for helpful discussions and advice, and D. Silver and J.-Y. Kim for their critical review of this manuscript. The pWZL expression vector and HMECs carrying a dominant-negative allele of the gene encoding p53 were generously provided by J. Zhao, (Dana-Farber Cancer Institute). This work was supported by the Breast Cancer Research Foundation in New York. Support also came from the US National Cancer Institute Specialized Program of Research Excellence in Breast Cancer at Harvard (CA89393) and a Department of Defense Concept Award (BC053041). The Trial of Principle trial was supported by the Fondation Luxembourgeoise contre le Cancer, by the Fonds National de la Recherche Scientifique (C.S., B.H.-K., C.D.), by the Brussels Region, by the Walloon Region (BIOWIN) and by the European Commission through the 'Advancing Clinico-Genomic Trials' project (FP6-2005-IST-026996). We thank Sanofi-Aventis for their support with adjuvant Taxotere, and we thank all of the participants in the Trial of Principle.
Author information
Authors and Affiliations
Contributions
A.L.R. and Z.C.W. designed the experiments and supervised the project. Yang Li performed the in vitro laboratory experiments including generating complementary DNA vector constructs, gene transfer and knockdown, RT-PCR and western blot analysis, and drug sensitivity and localization studies. L.Z. performed the PAM data analysis and statistics. Q.L., under supervision of Z.S., performed bioinformatic analysis on validation data sets and the cisplatin trial data set. R.T. performed apoptosis assays. Yan Li contributed to the preparation of cDNA vector constructs. C.D., C.S. and B.H.-K. performed the epirubicin trial and provided clinical data and analysis. A.L.R. provided the DF/HCC and cisplatin trial clinical samples and performed gene expression array analysis. Z.C.W. performed the SNP array analysis and scored the FISH assays. A.L.R., J.D.I. and Z.C.W. wrote the manuscript with comments from Yang Li, C.D., C.S. and Z.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 2–3 and Supplementary Methods (PDF 987 kb)
Supplementary Table 1
PAM probe list (XLS 38 kb)
Supplementary Table 4
Sample features (XLS 57 kb)
Rights and permissions
About this article
Cite this article
Li, Y., Zou, L., Li, Q. et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med 16, 214–218 (2010). https://doi.org/10.1038/nm.2090
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2090
This article is cited by
-
Autophagy modulators influence the content of important signalling molecules in PS-positive extracellular vesicles
Cell Communication and Signaling (2023)
-
High-throughput functional screen identifies YWHAZ as a key regulator of pancreatic cancer metastasis
Cell Death & Disease (2023)
-
miR-137–LAPTM4B regulates cytoskeleton organization and cancer metastasis via the RhoA-LIMK-Cofilin pathway in osteosarcoma
Oncogenesis (2023)
-
Loss of REST in breast cancer promotes tumor progression through estrogen sensitization, MMP24 and CEMIP overexpression
BMC Cancer (2022)
-
A critical ETV4/Twist1/Vimentin axis in Ha-RAS-induced aggressive breast cancer
Cancer Gene Therapy (2022)