Abstract
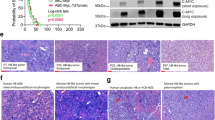
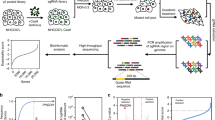
In solid tumors, resistance to therapy inevitably develops upon treatment with cytotoxic drugs or molecularly targeted therapies. Here, we describe a system that enables pooled shRNA screening directly in mouse hepatocellular carcinomas (HCC) in vivo to identify genes likely to be involved in therapy resistance. Using a focused shRNA library targeting genes located within focal genomic amplifications of human HCC, we screened for genes whose inhibition increased the therapeutic efficacy of the multikinase inhibitor sorafenib. Both shRNA-mediated and pharmacological silencing of Mapk14 (p38α) were found to sensitize mouse HCC to sorafenib therapy and prolong survival by abrogating Mapk14-dependent activation of Mek-Erk and Atf2 signaling. Elevated Mapk14-Atf2 signaling predicted poor response to sorafenib therapy in human HCC, and sorafenib resistance of p-Mapk14-expressing HCC cells could be reverted by silencing Mapk14. Our results suggest that a combination of sorafenib and Mapk14 blockade is a promising approach to overcoming therapy resistance of human HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Poulikakos, P.I. & Rosen, N. Mutant BRAF melanomas—dependence and resistance. Cancer Cell 19, 11–15 (2011).
Hartsough, E., Shao, Y. & Aplin, A.E. Resistance to RAF inhibitors revisited. J. Invest. Dermatol. 134, 319–325 (2014).
Berasain, C. Hepatocellular carcinoma and sorafenib: too many resistance mechanisms? Gut 62, 1674–1675 (2013).
Bruix, J., Gores, G.J. & Mazzaferro, V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut 63, 844–855 (2014).
Yau, T., Chan, P., Epstein, R. & Poon, R.T. Evolution of systemic therapy of advanced hepatocellular carcinoma. World J. Gastroenterol. 14, 6437–6441 (2008).
Lord, R., Suddle, A. & Ross, P.J. Emerging strategies in the treatment of advanced hepatocellular carcinoma: the role of targeted therapies. Int. J. Clin. Pract. 65, 182–188 (2011).
Llovet, J.M. et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 (2008).
Wilhelm, S. et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 5, 835–844 (2006).
Wuestefeld, T. et al. A direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell 153, 389–401 (2013).
Kang, T.W. et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011).
Carlson, C.M., Frandsen, J.L., Kirchhof, N., McIvor, R.S. & Largaespada, D.A. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl. Acad. Sci. USA 102, 17059–17064 (2005).
Braumüller, H. et al. T-helper-1-cell cytokines drive cancer into senescence. Nature 494, 361–365 (2013).
Holohan, C., Van, S.S., Longley, D.B. & Johnston, P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13, 714–726 (2013).
Margutti, S. & Laufer, S.A. Are MAP kinases drug targets? Yes, but difficult ones. ChemMedChem 2, 1116–1140 (2007).
Sawey, E.T. et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell 19, 347–358 (2011).
Wagner, E.F. & Nebreda, A.R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 9, 537–549 (2009).
Nebreda, A.R. & Porras, A. p38 MAP kinases: beyond the stress response. Trends Biochem. Sci. 25, 257–260 (2000).
Zhang, J., Shen, B. & Lin, A. Novel strategies for inhibition of the p38 MAPK pathway. Trends Pharmacol. Sci. 28, 286–295 (2007).
Goldstein, D.M., Kuglstatter, A., Lou, Y. & Soth, M.J. Selective p38alpha inhibitors clinically evaluated for the treatment of chronic inflammatory disorders. J. Med. Chem. 53, 2345–2353 (2010).
Dominguez, C., Powers, D.A. & Tamayo, N. p38 MAP kinase inhibitors: many are made, but few are chosen. Curr. Opin. Drug Discov. Devel. 8, 421–430 (2005).
Koeberle, S.C. et al. Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor. Nat. Chem. Biol. 8, 141–143 (2012).
Hope, H.R. et al. Anti-inflammatory properties of a novel N-phenyl pyridinone inhibitor of p38 mitogen-activated protein kinase: preclinical-to-clinical translation. J. Pharmacol. Exp. Ther. 331, 882–895 (2009).
Fischer, S. et al. Dibenzosuberones as p38 mitogen-activated protein kinase inhibitors with low ATP competitiveness and outstanding whole blood activity. J. Med. Chem. 56, 241–253 (2013).
MacNee, W., Allan, R.J., Jones, I., De Salvo, M.C. & Tan, L.F. Efficacy and safety of the oral p38 inhibitor PH-797804 in chronic obstructive pulmonary disease: a randomised clinical trial. Thorax 68, 738–745 (2013).
Calvisi, D.F. et al. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J. Hepatol. 54, 311–319 (2011).
Calvisi, D.F. et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 130, 1117–1128 (2006).
Guichard, C. et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44, 694–698 (2012).
Steelman, L.S. et al. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia 18, 189–218 (2004).
Hindley, A. & Kolch, W. Extracellular signal regulated kinase (ERK)/mitogen activated protein kinase (MAPK)-independent functions of Raf kinases. J. Cell Sci. 115, 1575–1581 (2002).
Turner, N.C. et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 27, 1368–1377 (2008).
Swanton, C. et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell 11, 498–512 (2007).
Giroux, V., Iovanna, J. & Dagorn, J.C. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. FASEB J. 20, 1982–1991 (2006).
Huang, S. et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell 151, 937–950 (2012).
Burgess, D.J. et al. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc. Natl. Acad. Sci. USA 105, 9053–9058 (2008).
Huesken, D. et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 23, 995–1001 (2005).
Mitchell, C. & Willenbring, H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3, 1167–1170 (2008).
Schlaeger, C. et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology 47, 511–520 (2008).
Dow, L.E. et al. A pipeline for the generation of shRNA transgenic mice. Nat. Protoc. 7, 374–393 (2012).
Acknowledgements
We thank A. Rinkel, N. Struever, C. Hermann, V. Geissler, U. Koppenhoefer, H. Riedesel, M. Jarek, M. Scharfe and the HZI Genome Analytics Group team for technical assistance. We thank the tissue bank of the National Center for Tumor Diseases Heidelberg for providing human HCC tissues. This work was supported by the German Research Foundation, DFG (Emmy Noether Programme ZE 545/2-1 to L.Z., the “Rebirth” Cluster of Excellence, project “Liver regeneration”, SFB/TRR77 and SFB685), the Helmholtz Association of German Research Centers (VH-NG-424 to L.Z.), the European Commission (project 'Heptromic') and the Wilhelm Sander Stiftung.
Author information
Authors and Affiliations
Contributions
The study was designed by L.Z. Research was conducted by R.R., D.D., T.L., K.M., T.W., T.-W.K., A.H., M.P., J.L., A.v.T., P.S., J.Z., K.-H.W., S.P., N.P.M., M.E., B.S., S.W.L., R.G., S.L. and L.Z. The manuscript was written by L.Z., R.R. and D.D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 (PDF 4565 kb)
Supplementary Table 1
Results from in vivo RNAi screen. (XLS 169 kb)
Rights and permissions
About this article
Cite this article
Rudalska, R., Dauch, D., Longerich, T. et al. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat Med 20, 1138–1146 (2014). https://doi.org/10.1038/nm.3679
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3679
This article is cited by
-
PLEKHG5 is stabilized by HDAC2-related deacetylation and confers sorafenib resistance in hepatocellular carcinoma
Cell Death Discovery (2023)
-
Mfap4: a promising target for enhanced liver regeneration and chronic liver disease treatment
npj Regenerative Medicine (2023)
-
Safety and efficacy of prophylactic and therapeutic vaccine based on live-attenuated Listeria monocytogenes in hepatobiliary cancers
Oncogene (2022)
-
Modelling liver cancer microenvironment using a novel 3D culture system
Scientific Reports (2022)
-
CRISPR-Cas9-based genome-wide screening identified novel targets for treating sorafenib-resistant hepatocellular carcinoma: a cross-talk between FGF21 and the NRF2 pathway
Science China Life Sciences (2022)