Abstract
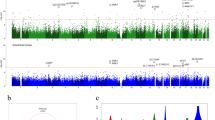
We used a collection of 708 prospectively collected autopsied brains to assess the methylation state of the brain's DNA in relation to Alzheimer's disease (AD). We found that the level of methylation at 71 of the 415,848 interrogated CpGs was significantly associated with the burden of AD pathology, including CpGs in the ABCA7 and BIN1 regions, which harbor known AD susceptibility variants. We validated 11 of the differentially methylated regions in an independent set of 117 subjects. Furthermore, we functionally validated these CpG associations and identified the nearby genes whose RNA expression was altered in AD: ANK1, CDH23, DIP2A, RHBDF2, RPL13, SERPINF1 and SERPINF2. Our analyses suggest that these DNA methylation changes may have a role in the onset of AD given that we observed them in presymptomatic subjects and that six of the validated genes connect to a known AD susceptibility gene network.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wang, S.C., Oelze, B. & Schumacher, A. Age-specific epigenetic drift in late-onset Alzheimer's disease. PLoS ONE 3, e2698 (2008).
Feinberg, A.P. et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci. Transl. Med. 2, 49ra67 (2010).
Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31, 142–147 (2013).
Chuang, D.M., Leng, Y., Marinova, Z., Kim, H.J. & Chiu, C.T. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 32, 591–601 (2009).
Gräff, J. et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 483, 222–226 (2012).
Chouliaras, L. et al. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol. Aging 34, 2091–2099 (2013).
Bakulski, K.M. et al. Genome-wide DNA methylation differences between late-onset Alzheimer's disease and cognitively normal controls in human frontal cortex. J. Alzheimers Dis. 29, 571–588 (2012).
Numata, S. et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 90, 260–272 (2012).
Akbarian, S., Beeri, M.S. & Haroutunian, V. Epigenetic determinants of healthy and diseased brain aging and cognition. JAMA Neurol. 70, 711–718 (2013).
Hernandez, D.G. et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 20, 1164–1172 (2011).
Bell, J.T. et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 8, e1002629 (2012).
Horvath, S. et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 13, R97 (2012).
Raj, T. et al. Alzheimer disease susceptibility loci: evidence for a protein network under natural selection. Am. J. Hum. Genet. 90, 720–726 (2012).
Bennett, D.A. et al. Selected findings from the Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers Dis. 33, S397–S403 (2013).
Bock, C. et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 28, 1106–1114 (2010).
Negash, S., Bennett, D.A., Wilson, R.S., Schneider, J.A. & Arnold, S.E. Cognition and neuropathology in aging: multidimensional perspectives from the Rush Religious Orders Study and Rush Memory And Aging Project. Curr. Alzheimer Res. 8, 336–340 (2011).
Bennett, D.A., Wilson, R.S., Boyle, P.A., Buchman, A.S. & Schneider, J.A. Relation of neuropathology to cognition in persons without cognitive impairment. Ann. Neurol. 72, 599–609 (2012).
Chibnik, L.B. et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann. Neurol. 69, 560–569 (2011).
Naj, A.C. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 43, 436–441 (2011).
Seshadri, S. et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. J. Am. Med. Assoc. 303, 1832–1840 (2010).
Hollingworth, P. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 43, 429–435 (2011).
Lambert, J.C. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 41, 1094–1099 (2009).
Shulman, J.M. et al. Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol. 70, 1150–1157 (2013).
Lunnon, K. et al. Cross-tissue methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer's disease neuropathology. Nat. Neurosci. 17, XXX–YYY (2014).
Schneider, J.A., Arvanitakis, Z., Leurgans, S.E. & Bennett, D.A. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann. Neurol. 66, 200–208 (2009).
Sperling, R.A. et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 7, 280–292 (2011).
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Chapuis, J. et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry 18, 1225–1234 (2013).
Lambert, J.C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458 (2013).
Tanaka, M. et al. DIP2 disco-interacting protein 2 homolog A (Drosophila) is a candidate receptor for follistatin-related protein/follistatin-like 1–analysis of their binding with TGF-beta superfamily proteins. FEBS J. 277, 4278–4289 (2010).
Cruchaga, C. et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature 505, 550–554 (2014).
Roet, K.C. et al. A multilevel screening strategy defines a molecular fingerprint of proregenerative olfactory ensheathing cells and identifies SCARB2, a protein that improves regenerative sprouting of injured sensory spinal axons. J. Neurosci. 33, 11116–11135 (2013).
Adrain, C., Zettl, M., Christova, Y., Taylor, N. & Freeman, M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science 335, 225–228 (2012).
Siggs, O.M. et al. iRhom2 is required for the secretion of mouse TNFalpha. Blood 119, 5769–5771 (2012).
McIlwain, D.R. et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science 335, 229–232 (2012).
Maretzky, T. et al. iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proc. Natl. Acad. Sci. USA 110, 11433–11438 (2013).
Cooper-Knock, J. et al. Gene expression profiling in human neurodegenerative disease. Nat. Rev. Neurol. 8, 518–530 (2012).
Simpson, J.E. et al. Microarray analysis of the astrocyte transcriptome in the aging brain: relationship to Alzheimer's pathology and APOE genotype. Neurobiol. Aging 32, 1795–1807 (2011).
Linnertz, C. et al. Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS ONE 4, e7480 (2009).
Renner, N.A. et al. Transient acidification and subsequent proinflammatory cytokine stimulation of astrocytes induce distinct activation phenotypes. J. Cell. Physiol. 228, 1284–1294 (2013).
Kraft, A.W. et al. Attenuating astrocyte activation accelerates plaque pathogenesis in APP/PS1 mice. FASEB J. 27, 187–198 (2013).
Bennett, D.A. et al. Overview and findings from the rush Memory and Aging Project. Curr. Alzheimer Res. 9, 646–663 (2012).
Bennett, D.A., Schneider, J.A., Arvanitakis, Z. & Wilson, R.S. Overview and findings from the religious orders study. Curr. Alzheimer Res. 9, 628–645 (2012).
Allen, M. et al. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology 79, 221–228 (2012).
Zou, F. et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 8, e1002707 (2012).
Du, P., Kibbe, W.A. & Lin, S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008).
Lin, S.M., Du, P., Huber, W. & Kibbe, W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 36, e11 (2008).
Bennett, D.A. et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 66, 1837–1844 (2006).
Chen, Y.A. et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209 (2013).
Johnson, W.E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Du, P. et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11, 587 (2010).
Guintivano, J., Aryee, M.J. & Kaminsky, Z.A. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 8, 290–302 (2013).
Bock, C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 13, 705–719 (2012).
Gifford, C.A. et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149–1163 (2013).
Xi, Y. & Li, W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009).
Ziller, M.J. et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481 (2013).
Zhu, J. et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642–654 (2013).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Rossin, E.J. et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7, e1001273 (2011).
Lage, K. et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25, 309–316 (2007).
Acknowledgements
We thank the National Institute for Health (NIHR) Biomedical Research Unit in Dementia in the South London and Maudsley NHS Foundation Trust (SLaM), Brains for Dementia Research (Alzheimer Brain Bank UK) and the donors and families who made this research possible. We also would like to thank the participants of the ROS and MAP studies for their participation in these studies. Support for this research was provided by grants from the US National Institutes of Health (R01 AG036042, R01AG036836, R01 AG17917,R01AG15819, R01 AG032990, R01 AG18023, RC2 AG036547, P30 AG10161, P50 AG016574, U01 ES017155, KL2 RR024151, K25 AG041906-01). Support was also provided by the Siragusa Foundation to N.E.-T., and the Robert and Clarice Smith and Abigail Van Buren Alzheimer's Disease Research Program to N.E.-T., S.G.Y. and F.Z. This work was funded by US National Institutes of Health grant AG036039 to J.M. and an Equipment Grant from Alzheimer's Research UK.
Author information
Authors and Affiliations
Contributions
C.M., A.T., W.B., S.G., C.B.E., B.E.B., A.M. and J.A.S. collected, prepared and generated data from the samples. G.S., L.B.C., J.E., B.T.K., M.K., T.R., J.R. and L.Y. performed analyses on the resulting data. K.L., L.C.S. and J.M. generated and analyzed the replication data. N.E.-T., J.B., H.S.C., C.Y., F.Z. and S.G.Y. provided and analyzed RNA data from AD and non-AD brains. P.L.D. and D.A.B. designed the study. P.L.D., D.A.B. and L.B.C. wrote the manuscript. All of the authors critically reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–7 and Supplementary Figures 1–6 (PDF 6819 kb)
Rights and permissions
About this article
Cite this article
De Jager, P., Srivastava, G., Lunnon, K. et al. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 17, 1156–1163 (2014). https://doi.org/10.1038/nn.3786
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3786
This article is cited by
-
The interaction between ageing and Alzheimer's disease: insights from the hallmarks of ageing
Translational Neurodegeneration (2024)
-
Multi-omics and pathway analyses of genome-wide associations implicate regulation and immunity in verbal declarative memory performance
Alzheimer's Research & Therapy (2024)
-
Longitudinal APOE4- and amyloid-dependent changes in the blood transcriptome in cognitively intact older adults
Alzheimer's Research & Therapy (2023)
-
Entorhinal cortex epigenome-wide association study highlights four novel loci showing differential methylation in Alzheimer’s disease
Alzheimer's Research & Therapy (2023)
-
The contribution of DNA methylation to the (dys)function of oligodendroglia in neurodegeneration
Acta Neuropathologica Communications (2023)