Abstract
Genetic variants conferring risk for autism spectrum disorder (ASD) have been identified, but the role of post-transcriptional mechanisms in ASD is not well understood. We performed genome-wide microRNA (miRNA) expression profiling in post-mortem brains from individuals with ASD and controls and identified miRNAs and co-regulated modules that were perturbed in ASD. Putative targets of these ASD-affected miRNAs were enriched for genes that have been implicated in ASD risk. We confirmed regulatory relationships between several miRNAs and their putative target mRNAs in primary human neural progenitors. These include hsa-miR-21-3p, a miRNA of unknown CNS function that is upregulated in ASD and that targets neuronal genes downregulated in ASD, and hsa_can_1002-m, a previously unknown, primate-specific miRNA that is downregulated in ASD and that regulates the epidermal growth factor receptor and fibroblast growth factor receptor signaling pathways involved in neural development and immune function. Our findings support a role for miRNA dysregulation in ASD pathophysiology and provide a rich data set and framework for future analyses of miRNAs in neuropsychiatric diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Bourgeron, T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 16, 551–563 (2015).
Geschwind, D.H. & State, M.W. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 14, 1109–1120 (2015).
Gaugler, T. et al. Most genetic risk for autism resides with common variation. Nat. Genet. 46, 881–885 (2014).
Geschwind, D.H. & Flint, J. Genetics and genomics of psychiatric disease. Science 349, 1489–1494 (2015).
Ha, M. & Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 (2014).
Friedman, R.C., Farh, K.K.H., Burge, C.B. & Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009).
Im, H.-I. & Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 35, 325–334 (2012).
Amaral, D.G., Schumann, C.M. & Nordahl, C.W. Neuroanatomy of autism. Trends Neurosci. 31, 137–145 (2008).
Friedländer, M.R., Mackowiak, S.D., Li, N., Chen, W. & Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52 (2012).
Friedländer, M.R. et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 15, R57 (2014).
Voineagu, I. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011).
Oldham, M.C. et al. Functional organization of the transcriptome in human brain. Nat. Neurosci. 11, 1271–1282 (2008).
Parikshak, N.N. et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013).
Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17 (2005).
Parikshak, N.N., Gandal, M.J. & Geschwind, D.H. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 16, 441–458 (2015).
Langfelder, P., Luo, R., Oldham, M.C. & Horvath, S. Is my network module preserved and reproducible? PLoS Comput. Biol. 7, e1001057 (2011).
Arbiza, L. et al. Genome-wide inference of natural selection on human transcription factor binding sites. Nat. Genet. 45, 723–729 (2013).
Ronan, J.L., Wu, W. & Crabtree, G.R. From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 14, 347–359 (2013).
Lewis, B.P., Burge, C.B. & Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Grimson, A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 (2007).
Garcia, D.M. et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18, 1139–1146 (2011).
Hsu, S.-D. et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 42, D78–D85 (2014).
Basu, S.N., Kollu, R. & Banerjee-Basu, S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 37, D832–D836 (2009).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Darnell, J.C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Bayés, A. et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 14, 19–21 (2011).
Kang, H.J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011).
Sanders, S.J. et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015).
Anney, R. et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum. Mol. Genet. 21, 4781–4792 (2012).
Wang, K. et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459, 528–533 (2009).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Lee, P.H., O'Dushlaine, C., Thomas, B. & Purcell, S.M. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics 28, 1797–1799 (2012).
Ripke, S. et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Lambert, J.-C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458 (2013).
Hirokawa, N., Niwa, S. & Tanaka, Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 (2010).
Lasiecka, Z.M. & Winckler, B. Mechanisms of polarized membrane trafficking in neurons -- focusing in on endosomes. Mol. Cell. Neurosci. 48, 278–287 (2011).
Wan, Q.-F. et al. SV2 acts via presynaptic calcium to regulate neurotransmitter release. Neuron 66, 884–895 (2010).
Redies, C., Hertel, N. & Hübner, C.A. Cadherins and neuropsychiatric disorders. Brain Res. 1470, 130–144 (2012).
Wong, R.W.C. & Guillaud, L. The role of epidermal growth factor and its receptors in mammalian CNS. Cytokine Growth Factor Rev. 15, 147–156 (2004).
Turner, C.A., Akil, H., Watson, S.J. & Evans, S.J. The fibroblast growth factor system and mood disorders. Biol. Psychiatry 59, 1128–1135 (2006).
Mellios, N. & Sur, M. The emerging role of microRNAs in schizophrenia and autism spectrum disorders. Front. Psychiatry 3, 39 (2012).
Rani, N. et al. A primate lncRNA mediates Notch signaling during neuronal development by sequestering miRNA. Neuron 90, 1174–1188 (2016).
Mundalil Vasu, M. et al. Serum microRNA profiles in children with autism. Mol. Autism 5, 40 (2014).
Mor, M., Nardone, S., Sams, D.S. & Elliott, E. Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Mol. Autism 6, 46 (2015).
Geschwind, D.H. & Rakic, P. Cortical evolution: judge the brain by its cover. Neuron 80, 633–647 (2013).
Geschwind, D.H. Advances in autism. Annu. Rev. Med. 60, 367–380 (2009).
Mariani, J. et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390 (2015).
Gawad, C., Koh, W. & Quake, S.R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016).
Quinn, E.M. et al. Development of strategies for SNP detection in RNA-seq data: application to lymphoblastoid cell lines and evaluation using 1000 Genomes data. PLoS One 8, e58815 (2013).
Friedländer, M.R. et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 26, 407–415 (2008).
Hackenberg, M., Sturm, M., Langenberger, D., Falcón-Pérez, J.M. & Aransay, A.M. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 37, W68–W76 (2009).
Hackenberg, M., Rodríguez-Ezpeleta, N. & Aransay, A.M. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 39, W132–W138 (2011).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Hansen, K.D., Irizarry, R.A. & Wu, Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13, 204–216 (2012).
Leek, J.T., Johnson, W.E., Parker, H.S., Jaffe, A.E. & Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Miller, J.A., Horvath, S. & Geschwind, D.H. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl. Acad. Sci. USA 107, 12698–12703 (2010).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Langfelder, P., Zhang, B. & Horvath, S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720 (2008).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal Complex Systems 1695 (2006).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Zambon, A.C., et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics 28, 2209–2210 (2012).
Stein, J.L. et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron 83, 69–86 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Li, H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Anders, S., Pyl, P.T. & Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Rossin, E.J. et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7, e1001273 (2011).
Lage, K. et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25, 309–316 (2007).
Acknowledgements
We would like to acknowledge the BrainSpan Consortium principal investigator N. Sestan (Yale University) for providing BrainSpan microRNA data and J. Ou for excellent technical assistance. This work was supported by US National Institutes of Health grant R01MH094714 to D.H.G. and is part of the PsychEncode Consortium.
Author information
Authors and Affiliations
Contributions
N.N.P. and T.G.B. performed brain sample dissections. Y.E.W. performed the other experiments and data analyses. N.N.P. provided code for differential gene expression, coexpression and enrichment analyses. D.H.G. provided guidance and oversight on all experiments and analyses. Y.E.W. and D.H.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
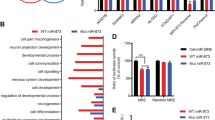
Supplementary Figure 1 Brain sample characteristics.
(a-b) Number of reads mapped to the hg19 reference genome (a) and mean read quality scores (b) for 131 ASD and 111 CTL brain tissue samples sequenced in this study. (c-e) Numbers of mapped reads, mean read quality scores, distribution of age, RIN, PMI, numbers of male and female individuals, numbers of prefrontal (FC) and temporal cortex (TC) samples, and numbers of samples from different brain banks (H, Harvard-ATP; N, NICHD-BTB) are shown for the ASD and CTL groups in 95 cortex samples used for the main DGE analysis (c), 109 cortex samples used for the main WGCNA analysis (d), and 47 cerebellum samples used for DGE analysis (e). Boxplot whiskers encompass data points within 1.5-fold interquartile ranges of the lower and upper quartiles.
Supplementary Figure 2 Principal component analysis and hierarchical sample clustering.
(a) Pearson correlations between different covariates and principal components (PCs) 1-5 of miRNA expression data (normalized for library size) for all 216 samples that passed quality control. Pearson correlation coefficients (R) and P values are indicated. Diagnosis (ASD vs. CTL), sex (female vs. male), and library preparation method (RiboZero selection vs. total RNA) were treated as binary numeric variables. Region (frontal cortex, temporal cortex, or cerebellum) and brain bank were treated as multi-level factor variables and adjusted R2 was calculated using a linear model between these two variables and other variables. Between region and brain bank, a chi-square test was performed and the P value was indicated in the heatmap (R was non-applicable). (b) Scatter plot of 216 samples based on PCs 1 and 2 of the miRNA expression data (normalized for library size). Dots are colored according to brain region. (c) Hierarchical clustering of all 216 samples using expression data (normalized for library size) of all expressed miRNAs. Information on diagnosis, age, sex, brain region, co-morbidity of seizures, psychiatric medication history, RIN, PMI, brain bank, and library preparation method is indicated with color bars below the dendrogram according to the legend on the right. (d) Pearson correlations between different covariates and PCs 1-5 of miRNA expression data (normalized for library size) for 95 cortex samples used for the main DGE analysis. Pearson correlation coefficients (R) and P values are indicated. Diagnosis (ASD vs. CTL), sex (female vs. male), region (frontal vs. temporal), and brain bank (Harvard-ATP vs. NICHD-BTB) were treated as binary numeric variables. (e) Scatter plot of 95 cortex samples based on PCs 1 and 2 of the miRNA expression data (normalized for library size). Dots are colored according to brain region. (f) Hierarchical clustering of 95 cortex samples using expression data (normalized for library size and other technical covariates) of all expressed miRNAs. Information on diagnosis, age, sex, brain region, co-morbidity of seizures, psychiatric medication history, RIN, PMI, and brain bank is indicated with color bars below the dendrogram according to the legend on the right.
Supplementary Figure 3 Robustness of DGE results.
(a) Comparison of miRNA fold changes in ASD vs. CTL between 10 rounds of random sampling of 70% of the samples and the original 95 cortex samples. Pearson correlation coefficients (R) and P values are indicated. Red line, y=x. (b-d) Comparison of miRNA fold changes in ASD vs. CTL between all 95 cortex samples and subsets of samples with RIN ≥ 5 (b), PMI ≤ 30 hrs (c), or no 15q duplication (d). Pearson correlation coefficients (R) and P values are indicated. Red line, y=x. (e) Comparison of miRNA fold changes in ASD vs. CTL between 95 cortex and 47 cerebellum samples. Red line, y=x; black line, linear regression between fold changes in the cerebellum and the cortex for miRNAs differentially expressed in the cortex (FDR < 0.05). Pearson’s R and P values are indicated. (f-g) Normalized log2(expression level) of down-regulated (f) and up-regulated (g) miRNAs in ASD (red) and control (blue) samples (n = 5 - 8) detected by qRT-PCR. Statistical significance was assessed using two-sided t-tests assuming unequal variance.
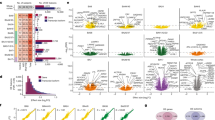
Supplementary Figure 4 Robustness, age dependence, and preservation of miRNA co-expression modules.
(a) Construction of consensus modules by bootstrapping for 200 rounds. Module assignment based on the original 109 samples, consensus module assignment based on 200 rounds of bootstrapping, and module assignment in each of the 200 resampled networks are shown below the dendrogram. The three ASD-associated modules are largely stable with perturbations to the initial subject and regional identity. The fraction of times each gene was assigned to the same module as in the consensus module is reported in Supplementary Table 2. (b-c) Comparison of ASD vs. CTL fold changes between samples from younger (15 - 30 years) and older (> 30 years) individuals for miRNAs in the yellow (b) and magenta (c) modules. Red line, y=x; black line, regression line between fold changes in the younger individuals and the older individuals for miRNAs differentially expressed (P < 0.05) in the younger set (orange and magenta dots). (d-g) Preservation of modules defined in ASD (d), CTL (e), TC (f), or FC (g) samples only in CTL (d), ASD (e), FC (f), or TC samples (g). (h-i) Module preservation analysis in two independent datasets: 31 independent cortex samples sequenced in this study (h) (Methods) and 167 samples covering 16 brain regions (neocortex, subcortical regions, thalamus, and cerebellum) and different ages (4 months to 19 years old) from the BrainSpan project (i). A module is considered not preserved if preservation Zsummary < 2, moderately preserved if 2 ≤ Zsummary < 10, and highly preserved if Zsummary ≥ 10.
Supplementary Figure 5 Experimental validation of TargetScan predicted miRNA targets.
Distribution (left) and cumulative distribution (right) of mRNA log2(fold change) in response to over-expression of hsa-miR-21-5p in hNPCs. Orange line, the strongest mRNA targets predicted by TargetScan (summed context+ score ≤ -0.1); purple line, the most conserved mRNA targets predicted by TargetScan (branch length in the top 25%, context+ score ≤ -0.05); red line, all mRNAs predicted to be hsa-miR-21-5p targets by TargetScan (the above two categories combined); green line, hsa-miR-21-5p targets documented in miRTarBase (452 miRNAs); black line, mRNAs not predicted to be hsa-miR-21-5p targets by TargetScan. Statistical significance between target groups and non-targets was assessed using ones-sided t-tests assuming unequal variance.
Supplementary Figure 6 Enrichment of ASD risk genes within the top targets of ASD-affected miRNAs and miRNA modules while controlling for 3’ UTR length.
(a) Heatmap showing enrichment of ASD SFARI genes, ID genes, ASD rare variants, FMRP targets, PSD genes, embryonically expressed genes, and chromatin modifiers, assessed using a logistic model that incorporates gene 3’ UTR length. P values were FDR corrected across 10 target groups for each gene list. (b) Heatmap showing enrichment of genes affected by multiple categories of de novo variants (DNVs), assessed using a logistic model that incorporates gene coding region length and gene 3’ UTR length. P values were FDR corrected across 10 target groups for each DNV category. (c) Heatmap showing enrichment of ASD-associated developmental gene co-expression modules in human cortex, assessed using a logistic model that incorporates gene 3’ UTR length. P values were FDR corrected across 10 target groups for each developmental module. Enrichment odds ratios (OR) and FDR corrected P values are shown for enrichments with FDR < 0.05.
Supplementary Figure 7 GO analysis for the predicted targets of differentially expressed miRNAs and ASD-related miRNA modules.
(a-b) Top relevant gene ontology categories (GO-Elite software, uncorrected P < 0.01, number of enriched genes > 5) for the strongest (a) or the most conserved (b) targets of the down-regulated and up-regulated miRNAs and the ASD-related miRNA modules. Enrichment Z scores represent relative enrichment in the targets compared to the background (Methods), with the red line at Z = 2. Asterisks indicate GO terms with FDR-corrected P values < 0.10.
Supplementary Figure 8 Effect of miRNAs on mRNA expression changes.
(a-h) Comparison of log2(fold change) distribution of down-regulated (a,b,e,f) or up-regulated (c,d,g,h) mRNAs (FDR < 0.05) that are predicted to be the strongest (a-d) or the most conserved (e-h) targets of the down-regulated or up-regulated miRNAs (a,c,e,g) or miRNA modules (b,d,f,h) to log2(fold change) distribution of differentially expressed mRNAs that are not predicted targets. The number of mRNAs in each group is indicated in brackets. One-tailed Wilcoxon rank sum tests were performed to compare each target group to the non-targets. *P < 0.05, **P < 0.01, ***P < 0.001. mRNAs that are predicted targets of both down-regulated and up-regulated miRNAs and miRNA modules were not included in the analysis.
Supplementary Figure 9 Correlation between differentially expressed miRNAs and mRNAs.
(a-c) Correlations between the PC1s of differentially expressed miRNAs (FDR < 0.05, |log2(fold change)| ≥ 0.3) and differentially expressed mRNAs (FDR < 0.05) that are predicted targets. (a) All differentially expressed miRNAs vs. all differentially expressed mRNAs; (b) up-regulated miRNAs vs. down-regulated mRNAs; (c) down-regulated miRNAs vs. up-regulated mRNAs. Pearson correlation coefficients (R) and P values within 47 CTL or 54 ASD samples alone, or the combined samples together are shown below the plots.
Supplementary Figure 10 Further characterization of hsa-miR-21-3p and hsa_can_1002-m.
(a) miRNA expression fold changes in the cortex plotted against percentile rank of mean expression levels across 95 cortex samples, with hsa-miR-21-3p and hsa_can_1002-m highlighted. (b-c) Views from the UCSC Genome Browser (https://genome.ucsc.edu) showing the chromosome locations and pre-miRNA sequences of hsa-miR-21-3p (b) and hsa_can_1002-m (c). The mature sequences and seed regions are indicated with black lines and green rectangles, respectively. Within the seed region of hsa_can_1002-m, a single-nucleotide difference indicated with a red asterisk was observed between the human and all other primate sequences. Gene annotations, multiple alignments of corresponding regions in different vertebrates, and measurements of evolutionary conservation using phyloP are shown. (d-g) Boxplots showing expression patterns of hsa-miR-21-3p (d,e) and hsa_can_1002-m (f,g) in different human brain regions (d,f) and at different developmental stages (e,g). DFC, dorsolateral prefrontal cortex; VFC, ventrolateral prefrontal cortex; MFC, medial prefrontal cortex; OFC, orbital frontal cortex; M1C, primary motor cortex; S1C primary somatosensory cortex; IPC, posteroinferior parietal cortex; A1C, primary auditory cortex; STC, posterior superior temporal cortex; ITC, inferolateral temporal cortex; V1C, primary visual cortex; HIP, hippocampus; AMY, amygdaloid complex; STR, striatum; MD, mediodorsal nucleus of thalamus; CBC, cerebellar cortex. Infancy, 4 months - 1 year; early childhood, 2 - 4 years; late childhood, 8 - 13 years; adolescence, 15 - 19 years; adulthood, 21 - 40 years. Boxplot whiskers indicate data points within 1.5-fold interquartile ranges of the lower and upper quartiles.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 3967 kb)
Supplementary Table 1
Brain sample metadata (XLSX 95 kb)
Supplementary Table 2
Information on predicted miRNAs, raw read counts, and DGE and WGCNA results (XLSX 1594 kb)
Supplementary Table 3
Enrichment of potential TF/CR targets in miRNA modules (XLSX 25 kb)
Supplementary Table 4
Numbers of miRNA targets and overlap between target groups (XLSX 41 kb)
Supplementary Table 5
Gene expression changes in hNPCs with miRNA over-expression (XLSX 21534 kb)
Supplementary Table 6
Enrichment analysis for miRNA targets (XLSX 107 kb)
Supplementary Code 1
Code for differential gene expression analysis using a linear mixed-effects model (TXT 3 kb)
Supplementary Code 2
Code for weighted gene co-expression network analysis using a bootstrapping-based method (TXT 7 kb)
Rights and permissions
About this article
Cite this article
Wu, Y., Parikshak, N., Belgard, T. et al. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat Neurosci 19, 1463–1476 (2016). https://doi.org/10.1038/nn.4373
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4373
This article is cited by
-
Exosome lncRNA IFNG-AS1 derived from mesenchymal stem cells of human adipose ameliorates neurogenesis and ASD-like behavior in BTBR mice
Journal of Nanobiotechnology (2024)
-
Exploring the molecular mechanism of comorbidity of autism spectrum disorder and inflammatory bowel disease by combining multiple data sets
Journal of Translational Medicine (2023)
-
Prenatal PM2.5 exposure impairs spatial learning and memory in male mice offspring: from transcriptional regulation to neuronal morphogenesis
Particle and Fibre Toxicology (2023)
-
Impact of IDO activation and alterations in the kynurenine pathway on hyperserotonemia, NAD+ production, and AhR activation in autism spectrum disorder
Translational Psychiatry (2023)
-
Integrative analysis of long noncoding RNAs dysregulation and synapse-associated ceRNA regulatory axes in autism
Translational Psychiatry (2023)