Abstract
Disrupted-in-Schizophrenia-1 (DISC1) is a genetic risk factor for a wide range of major mental disorders, including schizophrenia, major depression, and bipolar disorders. Recent reports suggest a potential role of DISC1 in the pathogenesis of Alzheimer’s disease (AD), by referring to an interaction between DISC1 and amyloid precursor protein (APP), and to an association of a single-nucleotide polymorphism in a DISC1 intron and late onset of AD. However, the function of DISC1 in AD remains unknown. In this study, decreased levels of DISC1 were observed in the cortex and hippocampus of 8-month-old APP/PS1 transgenic mice, an animal model of AD. Overexpression of DISC1 reduced, whereas knockdown of DISC1 increased protein levels, but not mRNA levels of β-site APP-Cleaving Enzyme 1 (BACE1), a key enzyme in amyloid-β (Aβ) generation. Reduction of BACE1 protein levels by overexpression of DISC1 was accompanied by an accelerating decline rate of BACE1, and was blocked by the lysosomal inhibitor chloroquine, rather than proteasome inhibitor MG-132. Moreover, overexpression of DISC1 in the hippocampus with an adeno-associated virus reduced the levels of BACE1, soluble Aβ40/42, amyloid plaque density, and rescued cognitive deficits of APP/PS1 transgenic mice. These results indicate that DISC1 attenuates Aβ generation and cognitive deficits of APP/PS1 transgenic mice through promoting lysosomal degradation of BACE1. Our findings provide new insights into the role of DISC1 in AD pathogenesis and link a potential function of DISC1 to the psychiatric symptoms of AD.
Similar content being viewed by others
Introduction
The amyloid hypothesis of Alzheimer’s disease (AD) maintains that the accumulation of amyloid-β (Aβ), especially its oligomeric forms, is a key event in disease pathogenesis (Masters and Beyreuther, 2006). The generation of Aβ is initiated by the proteolytic cleavage of amyloid precursor protein (APP) by β-secretase, which generates β-CTF, a C-terminal membrane-retained fragment of APP. The latter is further cleaved by γ-secretase and thus produces Aβ (Masters and Beyreuther, 2006). APP is also under sequential cleavage by α-secretase and γ-secretase, which precludes generation of Aβ (Sapra and Kim, 2009). Thus, β-secretase mediates the initial and rate-limiting processing step during Aβ generation. β-Site APP-cleaving enzyme 1 (BACE1), a type I transmembrane aspartyl protease, is the sole β-secretase in the brain. BACE1-deficient neurons do not produce detectable levels of Aβ40, Aβ42, or β-CTF, and AD mouse models with BACE1 deficiency do not generate amyloid deposition (Cai et al, 2001; Luo et al, 2001; Ohno et al, 2004). Moreover, protein levels and enzymatic activity of BACE1 are both increased in the brains of AD patients (Holsinger et al, 2002; Johnston et al, 2005; Yang et al, 2003). Therefore, inhibition of BACE1 is a potential therapeutic target for AD. Identification of molecules that reduce the increased levels/activity of BACE1 in the brains of AD patients may provide a worthwhile strategy in AD therapy.
Disrupted-in-Schizophrenia-1 (DISC1) is a genetic risk factor for a wide range of major mental disorders. Familial mutations in the DISC1 gene, including the balanced chromosomal (1; 11) (q42.1; q14.3) translocation, co-segregate with schizophrenia, major depression, and bipolar disorders (Schosser et al, 2010). This chromosomal translocation generates a C-terminal truncated or a fusion protein with DISC1 or a haploinsufficient DISC1 protein, which all cause dysfunctions ofDISC1 (Brandon and Sawa, 2011). DISC1 has essential roles in nervous system development, such as proliferation and differentiation of neural progenitor cells (Duan et al, 2007; Ishizuka et al, 2011; Lee et al, 2011), neuronal migration (Kamiya et al, 2005, 2008; Young-Pearse et al, 2010), neurite outgrowth (Miyoshi et al, 2003; Ozeki et al, 2003), synapse formation (Hayashi-Takagi et al, 2010; Kvajo et al, 2008; Lee et al, 2011), and mitochondrial trafficking (Atkin et al, 2011; Park et al, 2010). Interestingly, substantial evidence suggests that DISC1 is linked to AD. DISC1 interacts with APP (Young-Pearse et al, 2010), from which Aβ is derived by proteolytic cleavage through a BACE1-dependent mechanism. A genome-wide association study indicates an association of a single-nucleotide polymorphism in a DISC1 intron and late onset of AD (Beecham et al, 2009). However, the function of DISC1 in the pathogenesis of AD remains unknown. In our study, we observe decreased levels of DISC1 in the cortex and hippocampus of 8-month-old APP/PS1 transgenic mice, which express a chimeric mouse/human Swedish mutant APP (APPswe) and a mutant human presenilin 1 (PS1). We further provide evidence that DISC1 interacts with BACE1 and promotes lysosomal degradation of BACE1, thus reducing the generation of Aβ. Overexpression of DISC1 via an adeno-associated virus in the hippocampus reduces the levels of soluble Aβ40 and Aβ42, the density of Aβ plaques and, importantly, rescues cognitive deficits in APP/PS1 transgenic mice. Therefore, we propose that DISC1, a genetic risk factor for mental disorders, has essential roles in the pathology of AD.
Materials and Methods
Mice
APP/PS1 transgenic mice (stock number 004462) were purchased from the Jackson Laboratory and maintained by breeding with C57BL/6 mice. Littermates, matched in gender, were used in all experiments. Animal care and surgical procedures were approved by the Animal Studies Committee of Southern Medical University and of the Beijing Military Hospital in accordance with the international laws.
Antibodies
Anti-BACE1 (D10E5, CST, Danvers, MA); anti-DISC1 (Invitrogen, Grand Island, New York; Santa Cruz, Dallas, Texas); anti-Aβ (6E10, Covance, Dedham, MA); anti- lysosomal-associated membrane protein 1 (LAMP1) antibody (Abcam, Cambridge, MA); anti-APP and CTF (A8717), anti-Flag, anti-hemagglutinin (HA), and anti-β-actin were from Sigma-Aldrich (St Louis, MI); Alexa Fluor-conjugated secondary antibodies were from Invitrogen; horseradish peroxidase (HRP)-conjugated secondary antibodies were from Sigma-Aldrich.
Plasmids and siRNAs
Human full-length DISC1 cDNA containing a C-terminus-linked Flag was cloned into a pAOV vector containing the EF1a promoter, in which the green fluorescent protein (GFP) insert was deleted. A pAOV vector-expressing GFP served as control. Human BACE1 cDNA containing a C-terminus-linked HA was cloned into the pcDNA3.1 vector. DISC1 siRNAs and standard normal control (NC) siRNA were from Genepharma (Shanghai, China). Human DISC1 siRNA (sense: 5′-GGCAGAUGGAUGACUUAGATT-3′; anti-sense: 5′-UCUAAGUCAUCCAUCUGCCTT-3′); mouse DISC1 siRNA (sense: 5′-GGCAAACACUGUGAAGUGCTT-3′, antisense: 5′-GCACUUCACAGUGUUUGCCTT-3′); and NC siRNA (sense: 5′-UUCUCCGAACGUGUCACGUTT-3′; anti-sense: 5′-ACGUGACACGUUCGGAGAATT-3′) were used.
Cell Culture and Transfection
CHO cells and HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine. Lipofectamin 2000 (Invitrogen) and Effectene (Qiagen, Hilden, Germany) were used for transfection according to the manufacturer’s protocol. Primary neurons were cultured as described previously (Grunewald et al, 2012). Cortical and hippocampal neurons were prepared from embryonic day 18 (E18) C57 mice. Transfection of siRNA in cultured neurons was performed using Amaxa Nucleofector (Lonza) according to the instructions provided by the manufacturer. Neurons were then seeded in to six-well plates pre-coated with poly-D-lysine (Sigma) and cultured in Neurobasal medium supplemented with 2% B-27, 2 mM Glutamax-1 for 2 days.
Drug Treatment
MG-132 (M7449), chloroquine (C6628), and cycloheximide (C7698) were from Sigma-Aldrich. For measuring the time course of BACE1 degradation, HEK293 cells were treated with 40 μg/ml cycloheximide at 18 h after transfection of either DISC1-Flag or GFP. Cells were harvested at 0, 8, and 16 h after addition of cycloheximide. Cell lysates were collected at different time points after treatment and were then subjected to western blot analysis. For inhibition of lysosomal functions, HEK293 cells transfected with either DISC1-Flag or GFP were treated for 36 h with 25 μM chloroquine or 0.5 μM MG-132, lysed, and subjected to western blot analysis.
Reverse Transcription-PCR
Total cellular and tissue RNA were extracted using Trizol Reagent (Sigma-Aldrich). Equal amount of the first-strand cDNAs was synthesized with the FastQuant RT Kit (Tiangen Biotech, Beijing, China). PCR reactions were performed with Taq DNA polymerase (Takara, Dalian, China). The following primers were used: Human BACE1: 5′-ACCAACCTTCGTTTGCCCAA-3′ (forward), 5′-TCTCCTAGCCAGAAACCATCAG-3′ (reverse); Mouse Bace1: 5′-GTCATGAACGTAGCCCAGGT-3′ (forward), 5′-GAGAGGAATGTGAAGCCTCG-3′ (reverse); Human DISC1: 5′-GCACCCTGAGGAAGAAAGTT-3′ (forward), 5′-TCTCTTCTCAGTCTGTTGTAATCT-3′ (reverse); Human GAPDH: 5′-TTGGTATCGTGGAAGGACTCA-3′ (forward), 5′-TGTCATCATATTTGGCAGGTT-3′ (reverse). Mouse Gapdh: 5′-TCCACCACCCTGTTGCTGTAG-3′ (forward), 5′-GACCACAGTCCATGACATCACT-3′ (reverse).
Western Blot Analysis
Brain tissue and cells were lysed in RIPA buffer (50 mM Tris buffer (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing 1 mM PMSF and 1 × Protease Inhibitor Cocktail. Lysates were subjected to Tricine-SDS-PAGE for detection of CTFs, or SDS-PAGE for other proteins, followed by transferring to a polyvinylidene fluoride membrane (Merck Millipore, Darmstadt, Germany). The membranes were blocked with 5% BSA in Tris-buffered saline, pH 7.3 (TBS) containing 0.05% Tween-20 (TBST) for 1 h, and then incubated with primary antibodies diluted in blocking buffer for overnight at 4 °C. The membranes were washed with TBST and incubated with HRP-conjugated secondary antibodies (Sigma-Aldrich) for 2 h at room temperature. ECL Plus or ECL Advance Western Blotting Detection Reagents (Amersham Biosciences, Piscataway, NJ) were used to visualize the immunoreactive proteins. The intensities of the bands were quantified by Image J analysis (NIH, Bethesda).
Co-Immunoprecipitation
Mouse hippocampus and HEK293 cells were lysed with RIPA buffer containing the Protease Inhibitor Cocktail. The lysates were precleared with protein A/G plus agarose beads (Santa Cruz, Dallas) for 2 h and incubated together with antibody-conjugated beads overnight at 4 °C. The beads were washed three times with RIPA buffer, re-suspended in SDS electrophoresis sample buffer, and boiled for 5 min at 95 °C. Samples were subjected to SDS-PAGE and western blot analysis.
Virus Injection
Adeno-associated virus (serotype 8, AAV8) expressing human DISC1 or GFP were generated by Niuen Biotechnology (Shanghai, China). AAV8 viral particles (3.1 × 1012 vector genome/ml) were injected bilaterally into the hippocampus (−2.1 mm anteroposterior from bregma, ±1.8 mm mediolateral from bregma, and 1.8 mm below the surface of the skull) of 4-month-old APP/PS1 transgenic mice and gender-matched wild-type littermates (15 mice in each group) by a stereotaxic apparatus as described (Kiyota et al, 2010). AAV8-expressing GFP only was injected as control. One microliter of the virus suspension was injected into each hippocampus of a mouse through a 25-μl gauge needle at the rate of 0.2 μl/min. After injection, the needle was left in place for an additional 5 min before being slowly withdrawn. Mice were subjected to behavioral tests 3.5 months later and then killed for analysis of amyloid plaque deposition and tissue levels of Aβ and BACE1.
Morris Water Maze
This test was performed as described (Zhang et al, 2014). Briefly, mice were trained for 7 consecutive days, 4 trials per day. A different start location was used in each trial. Each trial lasted until the mice found the platform or for a 90 s. If the mice failed to find the platform in 90 s, they were gently guided to the platform and allowed to stay on the platform for 10 s. Escape latencies (time spent swimming from start point to the target) and path length (the distance from start point to the platform) before reaching the platform were recorded for 6 consecutive days. For the probe trial on day 7, the platform was removed. The numbers of times that the mice swam crossed the platform area were recorded during 60 s. In the reversal trial on day 8, the platform was moved to the opposite quadrant, and the mice received four training trials per day for 2 consecutive days. Escape latencies and the number of times that the mice swam across the platform area were recorded.
Immunofluorescence
Decapitated brains were immersed for fixation in 4% paraformaldehyde for 24 h followed by 30% sucrose in PBS, all at 4 °C. Coronal sections, 15 μm in thickness, were cut in a cryostat. Sections were washed three times with PBS containing 0.3% Triton X-100 (PBST), 10 min each, followed by blocking with 10% BSA in PBS for 1 h, and incubated with primary antibody overnight at 4 °C. Sections were washed three times in PBS and incubated with appropriate secondary Alexafluor-conjugated antibodies for 2 h at room temperature. The sections were then washed three times with PBS and mounted in mounting medium containing DAPI (Vector Laboratories, Burlingame). For immunocytochemistry, cells were fixed by 4% paraformaldehyde for 15 min. Cells were then washed and immersed for 0.1% Triton X-100 (PBST) for 5 min, followed by blocking with 10% BSA in PBS for 1 h, and incubated with primary antibody overnight at 4 °C. Cells were washed and incubated with appropriate secondary Alexa Fluor-conjugated antibodies for 1 h at room temperature.
ELISA for Measuring Levels of Aβ
For quantitation of Aβ, tissues from the cortex or hippocampus were lysed with solution A (5 M guanidine HCl, 50 mM Tris HCl, pH 8.0) and kept at room temperature for 3 h. The lysates were then diluted with cold Reaction Buffer B (Dulbecco’s phosphate-buffered saline, pH 7.4, containing 5% BSA, and 0.03% Tween-20) supplemented with Protease Inhibitor Cocktail to achieve optimal dilution, and centrifuged at 16 000 g for 20 min at 4 °C. The supernatant was then subjected to ELISA using the Aβ40 and Aβ42 ELISA KIT (Invitrogen) following the manufacturer’s instructions.
ELISA for Determining Proteins Interaction
BACE1 intracellular domain (a.a. 478–501) peptide was synthesized by China peptides Company (Shanghai, China). Purified recombinant DISC1 protein (transcript variant L) was purchased from Origene (Rockville, MD). 500 nM of BACE1 peptide (a.a. 478–501) in PBS was used for substrate coating in 96-well plates at 4 °C for overnight. The wells that were coated with PBS as substrate were used as the control. Wells were washed with PBS and blocked with 4% BSA in PBS for 1 h. Different concentrations of recombinant DISC1 protein (0, 5, 50, 100 nM) were added to each well and incubated for 1 h at 37 °C. Wells were then washed and incubated with mouse anti-DSIC1 antibody followed by incubation with HRP-conjugated goat anti-mouse IgG. OD values were read using the TMB system (Invitrogen).
Image Analysis and Aβ Plaque Quantification
Image analysis and Aβ plaque quantification were performed as described in Zhang et al (2014). Six mice per group were analyzed. Four sections of 15 μm in thickness from the hippocampus and cortex of each mouse were collected. All analyses were performed in corresponding sections from the same brain region in each group. Numbers and area of Aβ plaques of hippocampus and cortex in each image were quantified by Image J. The density of Aβ plaques was expressed as numbers of Aβ plaques per section. The size of Aβ plaques was quantified as the area occupied by Aβ plaques divided by the total area of the cortex or hippocampus.
Analysis of Translocation of BACE1 in Lysososmes
CHO cells were co-transfected with BACE1-HA and either DISC1 plasmid or the empty vector. Cells were treated with 25 μM chloroquine for 5 h at 16 h after transfection. Cells were then immunostained for HA, DISC1, and LAMP1. Images were acquired with a Zeiss confocal microscope. Image analysis and algorithm generation were performed using the Image-Pro Plus 6.0 software (Media Cybemetics, Silver Spring, MD). The Pearson’s correlation coefficient in each cell was calculated as previously described (Yu et al, 2006).
Statistical Analysis
All statistical analyses were performed using SPSS20.0. Data are presented as mean±SEM. Comparisons between two groups were made by independent samples t-test. Morris Water Maze data were measured by two-way repeated-measures ANOVA followed by the Fisher LSD test. p<0.05 was considered as significance level for all analyses. *p<0.05; **p <0.01; ***p<0.001.
Results
Decreased Levels of DISC1 in the Brains of APP/PS1 Transgenic Mice
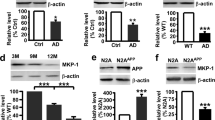
To investigate the potential role of DISC1 in the pathogenesis of AD, the levels of DISC1 in the hippocampus and cortex of APP/PS1 transgenic mice were investigated. Decreased levels of DISC1 were detected in the hippocampus (Figure 1a and b) and cortex (Figure 1c and d) of APP/PS1 transgenic mice at 8 months of age, when the mice display AD-like pathology, such as high load of amyloid plaques, loss of dendritic spines, and deficits in learning and memory (Zhang et al, 2014). In contrast, DISC1 exhibited identical levels in the hippocampus (Figure 1a and b) and cortex (Figure 1c and d) of APP/PS1 transgenic mice, in comparison to wild-type littermates, at 4 months of age, when the transgenic mice exhibit no cognitive deficits and harbor only a few amyloid plaques (Zhang et al, 2014). These results indicate a downregulation of DISC1 in the APP/PS1 brains, which may indicate a link to the pathogenesis of AD.
Decreased levels of DISC1 in the brains of APP/PS1 transgenic mice. (a and c) Western blot analysis of DISC1 in the hippocampus (a) and cortex (c) of APP/PS1 transgenic mice (TG) and of wild-type (WT) littermates at 4 and 8 months of age. Actin was used as loading control. (b and d) The levels of DISC1 in the hippocampus (b) and cortex (d) of wild-type mice were normalized to 1.0 for quantification of the relative levels of DISC1 in APP/PS1 transgenic hippocampus (b) or cortex (d). Data are presented as mean±SEM. **p<0.01. Independent samples t-test. n=3/group.
DISC1 Decreases Protein Levels of BACE1
In contrast to DISC1, levels of BACE1 have been reported to be increased in the APP/PS1 transgenic mouse brains at 10 months of age (Bao et al, 2013; Liang et al, 2010), suggesting a potential link between these two molecules. To further investigate the relationship between these two molecules, DISC1-Flag and BACE1-HA plasmids were co-transfected into HEK293 cells. The cells co-transfected with a plasmid expressing GFP and BACE1-HA plasmid were taken as control cells. Cell lysates were collected 48 h after transfection and subjected to western blot analysis with an antibody against HA (Figure 2a). Protein levels of BACE1 were decreased in DISC1-transfected cells compared with GFP-transfected control cells (Figure 2a and b). To further analyze whether DISC1 would downregulate endogenous BACE1 protein levels, HEK293 cells, which express BACE1 endogenously, were transfected with the DISC1-Flag plasmid, and then probed by western blot analysis with an antibody against BACE1 (Figure 2c). The protein levels of BACE1 were decreased in DISC1-transfected cells compared with those in GFP-transfected cells (Figure 2c and d). Thus, overexpression of DISC1 reduces levels of BACE1 in cultured cells. For further analysis, small interference RNA (siRNA) targeting DISC1 was transfected into HEK293 cells (Figure 2e–g) and primary cultured neurons derived from E18 mouse hippocampus and cortex (Figure 2h–j), which led to the downregulation of DISC1 levels in HEK293 cells (Figure 2e and g) and neurons (Figure 2h and j). The results showed that knockdown of DISC1 with siRNA-DISC1 enhanced protein levels of BACE1 in both HEK293 cells (Figure 2e and f) and neurons (Figure 2h and i). However, mRNA levels of BACE1 were similar in both DISC1-transfected (Figure 2k and l) and DISC1 siRNA-transfected HEK293 cells (Figure 2m and n), compared with control cells. These results indicate that DISC1 reduces BACE1 protein levels in a post-transcriptional manner.
DISC1 regulates the levels of BACE1 protein. (a) HEK293 cells were co-transfected with DISC-Flag and BACE1-HA plasmids or a plasmid in the same vector expressing GFP as control (GFP). The levels of BACE1 were probed with HA antibodies by western blot analysis. DISC1 was detected by Flag antibodies. Actin was used as loading control. (b) The levels of BACE1 in GFP-transfected cells were normalized to 1.0 for quantification of the relative levels of BACE1 in DISC1-transfected cells. (c) HEK293 cells were transfected with DISC1-Flag or GFP plasmid. The levels of BACE1 were probed with BACE1 antibodies by western blot analysis. DISC1 was detected by Flag antibodies. Actin was used as loading control. (d) Quantification of the band intensities in the western blots shown in c. (e and h) HEK293 cells (e) or primary cultured neurons derived from E18 mouse hippocampus (h) were transfected with DISC1 siRNA (siDISC1) or the scrambled siRNA (normal control, NC). The levels of BACE1 and DISC1 were probed with BACE1 and DISC1 antibodies by western blot analysis. Actin was used as loading control. (f, g, i and j) The levels of BACE1 and of DISC1 in NC-transfected cells were normalized to 1.0 for quantification of the relative levels of BACE1 (f and i) and of DISC1 (g and j) in DISC1-siRNA-transfected HEK293 cells (f and g) and DISC1-siRNA-transfected primary cultured hippocampal neurons (i and j). (k and m) The levels of BACE1 mRNA in HEK293 cells transfected with DISC1-Flag (k) or with DISC1-siRNA (siDISC1, m) were analyzed by reverse transcription-PCR. Actin was used as control. (l and n) The levels of BACE1 mRNA in the control cells were normalized to 1.0 for quantification of the relative levels of BACE1 mRNA. Data are presented as mean±SEM. *p<0.05; **p<0.01; ***p<0.001. Independent samples t-test. n=3/group.
DISC1 Associates with BACE1
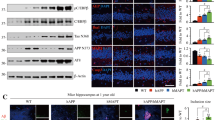
DISC1 acts as a cellular protein scaffold by interacting with various proteins and affects the function of different proteins (Bradshaw and Porteous, 2012). We further investigated whether DISC1 associates with BACE1. HEK293 cells co-transfected with DISC1-Flag and BACE1-HA were immunostained for DISC1 and BACE1 with antibodies against Flag and HA, respectively. As previously described, BACE1 immunoreactivity is detectable in perinuclear regions, which likely represent the Golgi apparatus and endoplasmic reticulum (Yan et al, 2001). DISC1 exhibited a staining pattern similar to BACE1 and colocalized with BACE1 in the perinuclear regions (Figure 3a). Co-immunoprecipitation was performed to investigate a potential interaction between these two molecules. Antibodies against HA (Figure 3b), Flag (Figure 3c) immunoprecipitated DISC1-Flag (Figure 3b), and BACE1-HA (Figure 3c), respectively, from HEK293 cells co-transfected with BACE1-HA and DISC1-Flag plasmids. Moreover, antibodies against BACE1 and DISC1 immunoprecipitated DISC1 and BACE1, respectively, from the lysates of adult mouse hippocampus (Figure 3d). To further analyze whether BACE1 directly binds to DISC1, ELISA experiment was performed. Using synthetic peptide that comprises the intracellular domain of BACE1 (a.a. 478–501) as the substrate coat, we observed a concentration-dependent binding of puried recombinant DISC1 protein to BACE1 peptide but not to the control (Figure 3e). Therefore, these results indicate that DISC1 binds to BACE1 in cells and the brains.
DISC1 associates with BACE1 and promotes degradation of BACE1 through lysosomes. (a) HEK293 cells co-transfected with BACE1-HA and DISC1-Flag were stained for Flag and HA. Scale bar: 25 μm. (b and c) HEK293 cells were co-transfected with BACE1-HA and DISC1-Flag. Cell lysates were collected 36 h after transfection and subjected to immunoprecipitation using HA antibody (b) or Flag antibody (c) and probed with Flag and HA antibodies, respectively. Cell lysates were loaded as input. (d) The lysates of hippocampus of 2-month-old C57BL/6J mice were used for immunoprecipitation with BACE1 or DISC1 antibodies and probed with DISC1 or BACE1 antibodies, respectively. Non-immune immunoglobulins (IgG) were used as control (mock). Tissue lysates were loaded as input. (e) The intracellular domain of BACE1 (a.a. 478–501) peptide binds to purified recombinant DISC1 in a concentration-dependent manner, as tested by ELISA. Binding was probed with anti-DISC1 antibody. (f) HEK293 cells transfected with either DISC1-Flag or GFP were treated with 40 μg/ml cycloheximide (CHX) for different time periods as indicated. Western blot analysis was performed with BACE1 and Flag antibodies. Actin was used as loading control. (g) The levels of BACE1 in either DISC1-Flag or GFP-transfected cells, which were collected before application of CHX treatment, were normalized to 1.0 for quantification of the relative levels of BACE1 in the cells treated with CHX for 8 and 16 h. (h) HEK293 cells transfected with DISC1-Flag or GFP were treated with either chloroquine (CQ), MG-132, or DMSO solution as vehicle control. Western blot analysis was performed with BACE1 and Flag antibodies. Actin was used as loading control. (i) The levels of BACE1 in vehicle-treated GFP-transfected cells were normalized to 1.0 for quantification of the relative levels of BACE1. (j) CHO cells co-transfected with BACE1-HA and DISC1 or empty vector were immnostained for DISC1, LAMP1, and HA, following treatment with chloroquine for 6 h to prevent lysosomal degradation of BACE1. (k) Analysis of the colocalization ratio between LAMP1± and BACE1-HA± fluorescent signal. Data are presented as mean±SEM. *p<0.05; **p<0.01; ***p<0.01. Independent samples t-test or one way ANOVA. n=3 or 4/group.
DISC1 Promotes Lysosomal Degradation of BACE1
As DISC1 decreases the levels of BACE1 in a post-transcriptional manner and that DISC1 associates with BACE1, we asked whether DISC1 may affect the degradation of BACE1. HEK293 cells were transfected with either DISC1-Flag or GFP plasmid. The transfected cells were treated with the protein synthesis inhibitor cycloheximide (Liu et al, 2005) at 18 h after transfection, when no significantly decreased levels of BACE1 had been yet observed by transfection of DISC1-Flag. The cell lysates were collected at 0, 8, and 16 h after addition of cycloheximide and subjected to western blot analysis with the BACE1 antibody. Levels of BACE1 in DISC1-Flag-transfected HEK293 cells were decreased after being treated by cycloheximide for 8 or 16 h, compared with those in GFP-transfected cells (Figure 3f). Normalization of BACE1 levels to 1.0 before cycloheximide treatment (0 h) in either DISC1-Flag or GFP-transfected cells were allowed to quantification of relative levels of BACE1 after addition of cycloheximide for 8 and 16 h (Figure 3g). BACE1 levels in DISC1-transfected HEK293 cells declined at a faster rate compared with those in GFP-transfected cells (Figure 3g). These results suggest that DISC1 promotes the degradation of BACE1 protein, which had been reported to occur via lysosomes (Koh et al, 2005). To examine this possibility, HEK293 cells were transfected with either DISC1-Flag or GFP and treated with the lysosomal inhibitor chloroquine or the proteasome inhibitor MG-132 (Koh et al, 2005) immediately after transfecion (Figure 3h). The downregulation of BACE1 levels by transfection with DISC-Flag was blocked by treatment with chloroquine, but not with MG-132 (Figure 3h and i). These results indicate that DISC1 promotes degradation of BACE1 through lysosomes. Considering that DISC1 has a role in regulation of the intracellular trafficking of protein and the fact that BACE1 is trafficked between subcellular compartments, such as the trans-Golgi network (TGN), the cell surface, and endosomes, and is eventually degraded in the lysosome (Jiang et al, 2014; Vassar et al, 2009), we examined whether DISC1 promotes translocation of BACE1 to lysosomes. CHO cells co-transfected with BACE1-HA and DISC1 were immunostained for HA and LAMP1, the marker of the lysosome (Figure 3j). BACE1 is degraded in the lysosome, which makes very few BACE1 be detected in the lysosome (Kandalepas et al, 2013; Koh et al, 2005). Thus, in this experiment, we treated transfected CHO cells with chloroquine for 6 h to prevent lysosomal degradation of BACE1 before immnostaining. The results showed that enhanced colocalization ratio between BACE1 and LAMP1 in DISC1-transfected cells compared with that in vector-transfected control cells (Figure 3k), indicating that overexpression of DISC1 promotes translocation of BACE1 into lysosomes.
Overexpression of DISC1 Reduces Expression of BACE1 in APP/PS1 Transgenic Mice
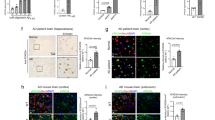
DISC1 reduces BACE1 protein levels through promoting lysosomal degradation of BACE1 in cultured cells. We then further validated whether DISC1 regulates BACE1 expression in the mouse brain. We injected an adeno-associated virus (serotype 8, AAV8) encoding DISC1-Flag into the hippocampus of APP/PS1 transgenic mice at 4 months of age. AAV8 expressing GFP was injected as control (Figure 4a and b). The expression of DISC1 via the virus was shown in the hippocampus by immunostaining for Flag (Figure 4c) 4 months after injection. Expression of DISC1 was further confirmed by western blot analysis (Figure 4d). A band at ~100 kDa corresponding to the expression of DISC1-Flag in the hippocampus was detected by Flag antibody (Figure 4d). Consistent with the results from cultured HEK293 cells, levels of BACE1 decreased in DISC1-AAV8-injected, compared with those in GFP-AAV8-injected hippocampus (Figure 4d and e). In contrast, BACE1 mRNA levels were similar in DISC1-Flag-AAV8-injected and GFP-AAV8-injected hippocampus (Figure 4f and g). These results indicate that overexpression of DISC1 reduces the levels of BACE1 protein in the brain.
Overexpression of DISC1 in the hippocampus of APP/PS1 transgenic mice reduces levels of BACE1 and cleavage of APP by BACE1. Hippocampus of APP/PS1 transgenic mice were injected with AAV8 encoding DISC1-Flag (DISC1) or GFP (GFP) and mice were maintained for 4 months before killing. (a–c) Coronal sections of the hippocampus of GFP-injected (a and b) or AAV-DISC1-Flag-injected (c) transgenic mice were stained for GFP (a and b) or Flag (c). GFP fluorescence in serial sections of the hippocampus is shown in a. Scale bars: 100 μm in the images at lower magnification, 25 μm in the images at higher magnification. (d) Western blot analysis of the lysates of hippocampus of transgenic mice injected with DISC1-Flag-AAV8 or GFP-AAV8 using BACE1, Flag, and actin antibodies. (e) The levels of BACE1 protein in the hippocampus of GFP injected APP/PS1 mice were normalized to 1.0 for quantification of the relative levels of BACE1 in DISC1-Flag-injected hippocampus. (f) Reverse transcription-PCR analysis of the levels of BACE1 mRNA and transfected human DISC1 mRNA in the hippocampus of transgenic mice injected with DISC1-Flag or GFP AAV8. Actin was used as loading control. (g) The levels of BACE1 mRNA in the hippocampus of GFP-injected transgenic mice were normalized to 1.0 for quantification of the relative levels of BACE1 mRNA in DISC1-Flag-injected hippocampus. (h) Western blot analysis of the levels of APP, β-CTF, α-CTF in the hippocampus of transgenic mice injected with AAV8 encoding GFP or DISC1-Flag with an antibody against the intracellular domain of APP. (i–k) The levels of β-CTF (i), α-CTF (j), and APP (k) in the hippocampus of GFP-injected mice were normalized to 1.0 for quantification their relative levels in DISC1-injected mice. Data are presented as mean±SEM. *p<0.05; **p<0.01. Independent samples t-test. n=3 or 4/group.
Overexpression of DISC1 Reduces Cleavage of APP by BACE1 in APP/PS1 Transgenic Mice
As DISC1 promotes the degradation of BACE1, which is the β-secretase in the brain, we investigated whether overexpression of DISC1 in the hippocampus would affect the cleavage of APP by BACE1 and thus generation of Aβ. Expression of β- or α-CTF, the C-terminal fragment of APP after being cleaved by BACE1 or α-secretase, respectively, were examined by western blot with an antibody against the intracellular domain of APP. β-CTF exhibited decreased levels in the hippocampus of DISC1-AAV8-injected APP/PS1 transgenic mice at 8 months of age, ie, 4 months after injection of virus, in comparison to transgenic mice injected with GFP-AAV8 (Figure 4h and i). In contrast, α-CTF (Figure 4h and j) and full-length APP (Figure 4h and k) were found at similar levels in the hippocampus of DISC-injected and GFP-injected APP/PS1 mice. These results indicate that DISC1 decreases the cleavage of APP by BACE1 rather than by α-secretase.
Overexpression of DISC1 Reduces the Density of Amyloid Plaques in APP/PS1 Transgenic Mice
Consistent with the results that DISC1 decreases BACE1-cleavage of APP, a key step for Aβ generation, decreased levels of soluble Aβ40 (Figure 5a), and Aβ42 (Figure 5b) were detected by ELISA in the hippocampus, but not in the cortex of DISC1-injected APP/PS1 mice. The ratio of Aβ40/Aβ42 levels were similar in the hippocampus of DISC1-injected and GFP-injected APP/PS1 mice (Figure 5c), suggesting that DISC1 may have minimal effect on γ-secretase cleavage of CTFs (Yin et al, 2007). Moreover, amyloid plaques exhibited reduced numbers (Figure 5d, e, g and h) and smaller size (Figure 5d, f, g and i) in the hippocampus (Figure 5d and f), but not in the cortex (Figure 5g and i), of DISC1-injected compared with those in GFP-injected APP/PS1 transgenic mice. These results indicate that overexpression of DISC1 reduces the generation of Aβ and thus the density of amyloid plaques in APP/PS1 transgenic mice.
Overexpression of DISC1 in the hippocampus of APP/PS1 transgenic mice reduces Aβ generation and rescues cognitive deficits of APP/PS1 transgenic mice. APP/PS1 transgenic mice were injected with AAV8 encoding DISC1-Flag (DISC1) or GFP (GFP) at 4 months of age and were maintained for 3.5–4 months before killing. (a and b) ELISA analysis of the levels of Aβ40 (a) and Aβ42 (b) in the hippocampus and cortex of transgenic mice injected with AAV8 encoding GFP or DISC1-Flag. (c) Analysis of the ratio of the levels of Aβ40/Aβ42 in the hippocampus of transgenic mice injected with AAV8 encoding GFP or DISC1-Flag. (d-i) Coronal sections of hippocampus (d) and cortex (g) of transgenic mice 4 months after injection of AAV8 encoding GFP or DISC1-Flag were immunostained for Aβ and counterstained with DAPI. The number (e and h) and the size (f and i) of amyloid plaques in the hippocampus (d–f) and cortex (g–i) of virus-injected transgenic mice were quantified. (j–p) Morris Water Maze test. Escape latencies (j), average swimming speeds (k), representative images of swimming paths (l), distances swum (m), number of platform site crossings in the probe trials (n), escape latencies (o), and number of platform site crossings in the reversal trials (p) are shown. (q) Schematic summary of the binding partners of DISC1 related to AD pathogenesis and their roles in Aβ generation. Interaction of DISC1 and APP promotes endocytosis of APP to the endosome, and thus enhances Aβ generation. Whereas binding of DISC1 to BACE1 promotes translocation of BACE1 to the lysosome, and thus accelerates its degradation. Data are presented as mean+SEM. *p<0.05; ***p<0.001. Independent samples t-test (a–i). n=6/group (a–i). Two-way repeated-measures ANOVA followed by the Fisher LSD test (j–p). n=6–13/group (j–p).
Overexpression of DISC1 Rescues Cognitive Deficits of APP/PS1 Transgenic Mice
As overexpression of DISC1 decreases the density of amyloid plaques, which cause cognitive deficits in APP/PS1 transgenic mice, we examined whether overexpression of DISC1 would rescue the deficits in learning and memory of these mice. AAV8 encoding either DISC1-Flag or GFP was injected into the hippocampus of 4-month-old transgenic mice, which were then subjected to Morris water maze test 3.5 months after virus injection. Wild-type littermates injected into the hippocampus with AAV8 encoding GFP were used as controls. As described (Zhang et al, 2014), GFP-injected transgenic mice were impaired in learning to use the available visuospatial cues to locate the submerged platform, as indicated by their longer escape latency compared with AAV8-GFP-injected wild-type mice (Figure 5j). As expected, the escape latency of DISC1-injected transgenic mice was reduced compared with that of GFP-injected transgenic mice, and even approached a level comparable to GFP-injected wild-type mice (Figure 5j). The changes in escape latency were not due to the differences in swimming speed, which were identical in the groups (Figure 5k). Consistently, DISC1-overexpressing transgenic mice exhibited shorter swimming paths to locate the target, when compared with GFP-injected transgenic mice (Figure 5l and m). After removal of the platform, although there is no statistic difference, a trend of increased number of times that DISC1-overexpressing transgenic mice swam across the platform area was observed, compared with that in GFP-injected transgenic mice (Figure 5n). In the reversal trials, where the platform was moved to the opposite quadrant, DISC1-injected transgenic mice also exhibited shorter escape latencie (Figure 5o) and increased number of times that swam cross the platform area (Figure 5p), which are comparable to those of GFP-injected wild-type mice. These results indicate that overexpression of DISC1 in the hippocampus rescues cognitive deficits of APP/PS1 transgenic mice.
Discussion
DISC1 is well known as a genetic risk factor for mental disorders, such as schizophrenia, major depression, and bipolar disorder. The present study reveals a novel role of DISC1 in AD involving an interaction with BACE1. DISC1 reduces the stability of BACE1 through interaction with BACE1 in a lysosome-dependent mechanism. DISC1 has been reported involved in modulation of the stability of serine racemase (SR) by promoting the ubiquitination-dependent degradation in the proteasome in astrocytes (Ma et al, 2013). However, the effect of DISC1 on the stability of SR is astrocyte specific. SR levels remain unchanged in DISC1 mutant neurons (Ma et al, 2013). BACE1 is predominantly expressed in neurons rather than in astrocytes (Vassar et al, 1999). These results suggest that DISC1 modulates the stability of certain proteins through different mechanisms in neurons and astrocytes. DISC1 acts as a cellular protein scaffold by interacting with and affecting the function of different proteins at different locations and times in development (Bradshaw and Porteous, 2012). For example, DISC1 interacts with APP and reduces expression of APP on the cell surface via enhancing the internalization of APP (Shahani et al, 2015; Figure 5q). DISC1 interacts with BACE1, which is trafficked between subcellular compartments, such as the TGN, the cell surface, and endosomes to be eventually degraded in the lysosome (Jiang et al, 2014; Vassar et al, 2009), and promotes translocation of BACE1 to lysosomes, and thus accelerates its degradation (Figure 5q).
DISC1 shows reduced levels in APP/PS1 brains and promotes BACE1 degradation, being in agreement with the observations on elevated levels of BACE1 in the brains of AD patients (Holsinger et al, 2002; Johnston et al, 2005; Yang et al, 2003) and APP/PS1 transgenic mice (Bao et al, 2013; Liang et al, 2010). It remains to be shown whether reduced expression of DISC1 occurs in the brains of AD patients and whether such a reduction would be correlated with levels of BACE1. However, it should be noted that BACE1 regulates the transcription of a subtype of DISC1 through cleavage of neuregulin, which binds to ErB4 after being cleaved (Seshadri et al, 2010). Thus, a feedback loop appears to exist between BACE1 and DISC1. Overexpression of DISC1 reduces protein levels of BACE1, whereas the reduction of BACE1 levels diminishes DISC1 transcription. Such a feedback loop may be a complementary mechanism in the maintenance of physiological expression of DISC1 and BACE1 in the brain. However, it is still unknown which factors disrupt such balance that eventually leads to reduced expression of DISC1 protein in the presence of elevated levels of BACE1. Further investigation on mRNA levels of DISC1 or epigenetic regulation of DISC1 in the brains of both AD patients and AD transgenic mice at different pathologic stages may help to answer this question.
A recent study has reported that knockdown or knockout of DISC1 inhibits Aβ generation through promoting expression of APP on the cell surface (Shahani et al, 2015), where α-secretase primarily resides (Sisodia, 1992). In contrast, we have observed in the present study that overexpression of DISC1 inhibits Aβ generation through reducing levels of BACE1, which tends to be catalytically active in the environment of the TGN or endosomal-lysosomal compartments (Sathya et al, 2012). Both studies have observed that DISC1 does not change the levels of full-length APP. It is thus possible that enough full-length APP is available for being cleaved by BACE1, whereas some portion of APP remains on the cell surface undergoing α-secretase cleavage. This idea is consistent with the fact that levels of β-CTF are decreased by overexpression of DISC1 in the hippocampus of APP/PS1 transgenic mice as shown in the present study and are enhanced by knockdown or knockout of DISC1 as revealed by the shown western blots in a previous study (Shahani et al, 2015). Levels of α-CTF are shown as a trend of decrease, but not statistically, by overexpression of DISC1 in this study and are increased significantly by knockdown or knockout of DISC1 in finding of (Shahani et al (2015). Considering that β-CTF could be cleaved by α-secretase on the cell surface (Jager et al, 2009), the cleavage of APP by α- and β-secretase might not always inversely correlate with each other as originally thought (Colombo et al, 2013; Wang et al, 2013). Therefore, it is possible that DISC1 reduces both α- and β-secretase cleavage of APP via increasing internalization of APP and promoting lysosomal degradation of BACE1, respectively.
It has been suggested that wild-type APP (APPwt) and Swedish mutant APP (APPswe) utilize different cellular mechanisms for Aβ generation. β-Secretase cleavage of APPwt occurs in endocytic compartments after internalization from the cell surface (Jager et al, 2009), whereas β-secretase cleavage of APPswe occurs late in the secretory pathway, which is almost independent of a functional internalization signal (Citron et al, 1992, 1995; Haass et al, 1995; Thinakaran et al, 1996; Jager et al, 2009). In this study, overexpression of DISC1 reduces Aβ generation in APP/PS1 transgenic mice that harbor APP Swedish mutations (APP695 K595N/M596L) and a human PS1 mutation (deletion of exon 9) (Reiserer et al, 2007). Thus, on the one hand, the APPswe mutation provides a preference of cleavage of APP by β-secretase (Citron et al, 1992). On the other hand, β-secretase cleavage of APPswe is independent of APP internalization, whereas internalization of endogenous APPwt is increased by overexpression of DISC1 (Citron et al, 1992, 1995; Shahani et al, 2015). These combined observations suggest that BACE1, rather than α-secretase, is the predominant secretase that cleaves APP in the hippocampus of APP/PS1 transgenic mice overexpressing DISC1. Thus, in this case, overexpression of DISC1 reduces Aβ generation in the presence of reduced levels of BACE1. Knockdown of DISC1 increases cell surface expression of APPwt by preventing its internalization (Shahani et al, 2015). APP and α/β-CTF share a NPTY motif in the cytoplasmic domain, which represents a consensus sequence for the internalization of transmembrane cell surface proteins (Chen et al, 1990; Jager et al, 2009). It is therefore possible that knockdown of DISC1 promotes cell surface expression of both full-length APP and α/β-CTF, which may reduce Aβ generation with the view that β-CTF at the cell surface could be further cleaved by α-secretase (Jager et al, 2009). Thus, the discrepancy in Aβ generation between our study and study by Shahani et al. may due to the different extents in dependency on internalization by BACE1-cleavage of APPwt and APPswe and the methodological differences in overexpression and knockdown system. The paradoxical finding in Aβ generation in this study and in study Shahani et al (2015) is also consistent with that high BACE1 overexpression in an AD transgenic model decreases Aβ generation and thus the density of amyloid plaques, whereas moderate or low BACE1 overexpression increases Aβ generation (Lee et al, 2005). Thus, more investigation, eg, overexpression of DISC1 in distinct AD mouse models or crossbreeding DISC1 knockout mice with APP/PS1 transgenic mice, will help to address this question.
It is noteworthy that the observations on reduction of Aβ generation by overexpression of DISC1 in the hippocampus are paralleled by the rescue of cognitive deficits of APP/PS1 transgenic mice, indicating that amelioration of plaque generation via DISC1 reducing levels of the Aβ-generating enzyme BACE1 ameliorates cognitive deficits in an animal model of AD. These observations further highlight that downregulation of DISC1 expression may causally contribute to cognitive deficits of APP/PS1 mice. Consistently, mice expressing mutant DISC1 show deficits in working memory and short-term synaptic plasticity (Koike et al, 2006; Kvajo et al, 2008, 2011). Both APP and BACE1, two binding partners of DISC1 (Young-Pearse et al, 2010), modulate synaptic plasticity (Filser et al, 2015; Petrus and Lee, 2014). In contrast to that DISC1 promotes internalization of APP from the cell surface (Shahani et al, 2015), APP also shows a role in promoting the location of DISC1 in the centrosome (Young-Pearse et al, 2010), and thus regulates neuronal migration, a process essential in maintenance of synaptic plasticity in the brain. It has been reported that regulation of neuronal migration by APP requires full-length APP (Young-Pearse et al, 2010). Our study shows that DISC1 in the cytoplasm accelerates degradation of BACE1 through promoting its translocation in the lysosome. Thus, it is possible that decreased levels of DISC1 enhances the cleavage of APP by BACE1 and thus decreases the levels of full-length APP, which is required in regulation of neuronal migration. However, it requires more experiments to address this possibility. It also requires further investigation on whether such process contributes to cognitive deficits of APP/PS1 mice independent on Aβ.
Moreover, several neuropsychiatric symptoms, such as abnormalities in emotionality/activity, social interactions, and sensomotor abilities are commonly detected in AD patients (Lyketsos et al, 2011). Deficits in working memory and social interactions that have been described for DISC1 mutant mice (Ayhan et al, 2011; Koike et al, 2006; Pletnikov et al, 2008) are also observed in BACE1 knock-in transgenic mice (Plucinska et al, 2014). Thus, the present study may suggest a possibility that decreased levels of DISC1 may account for the neuropsychiatric symptoms in AD. Investigations of DISC1 in relation to the neuropsychiatric symptoms of AD will not only suggest a basis for understanding these aspects of AD, but hopefully also help to understand some aspects of other mental diseases.
Funding and Disclosure
The authors declare no conflict of interest.
References
Atkin TA, MacAskill AF, Brandon NJ, Kittler JT (2011). Disrupted in Schizophrenia-1 regulates intracellular trafficking of mitochondria in neurons. Mol Psychiatry 16: 122–124.
Ayhan Y, Abazyan B, Nomura J, Kim R, Ladenheim B, Krasnova IN et al (2011). Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Mol Psychiatry 16: 293–306.
Bao XQ, Li N, Wang T, Kong XC, Tai WJ, Sun H et al (2013). FLZ alleviates the memory deficits in transgenic mouse model of Alzheimer's disease via decreasing beta-amyloid production and tau hyperphosphorylation. PLoS ONE 8: e78033.
Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL et al (2009). Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet 84: 35–43.
Bradshaw NJ, Porteous DJ (2012). DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology 62: 1230–1241.
Brandon NJ, Sawa A (2011). Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci 12: 707–722.
Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL et al (2001). BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci 4: 233–234.
Chen WJ, Goldstein JL, Brown MS (1990). NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem 265: 3116–3123.
Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P et al (1992). Mutation of the beta-amyloid precursor protein in familial Alzheimer's disease increases beta-protein production. Nature 360: 672–674.
Citron M, Teplow DB, Selkoe DJ (1995). Generation of amyloid beta protein from its precursor is sequence specific. Neuron 14: 661–670.
Colombo A, Wang H, Kuhn PH, Page R, Kremmer E, Dempsey PJ et al (2013). Constitutive alpha- and beta-secretase cleavages of the amyloid precursor protein are partially coupled in neurons, but not in frequently used cell lines. Neurobiol Dis 49: 137–147.
Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y et al (2007). Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130: 1146–1158.
Filser S, Ovsepian SV, Masana M, Blazquez-Llorca L, Brandt Elvang A, Volbracht C et al (2015). Pharmacological Inhibition of BACE1 Impairs Synaptic Plasticity and Cognitive Functions. Biol Psychiatry 77: 729–739.
Grunewald E, Tew KD, Porteous DJ, Thomson PA (2012). Developmental expression of orphan G protein-coupled receptor 50 in the mouse brain. ACS Chem Neurosci 3: 459–472.
Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D et al (1995). The Swedish mutation causes early-onset Alzheimer's disease by beta-secretase cleavage within the secretory pathway. Nat Med 1: 1291–1296.
Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ et al (2010). Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci 13: 327–332.
Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G (2002). Increased expression of the amyloid precursor beta-secretase in Alzheimer's disease. Ann Neurol 51: 783–786.
Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF et al (2011). DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature 473: 92–96.
Jager S, Leuchtenberger S, Martin A, Czirr E, Wesselowski J, Dieckmann M et al (2009). alpha-secretase mediated conversion of the amyloid precursor protein derived membrane stub C99 to C83 limits Abeta generation. J Neurochem 111: 1369–1382.
Jiang S, Li Y, Zhang X, Bu G, Xu H, Zhang YW (2014). Trafficking regulation of proteins in Alzheimer's disease. Mol Neurodegener 9: 6.
Johnston JA, Liu WW, Todd SA, Coulson DT, Murphy S, Irvine GB et al (2005). Expression and activity of beta-site amyloid precursor protein cleaving enzyme in Alzheimer's disease. Biochem Soc Trans 33: 1096–1100.
Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y et al (2005). A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol 7: 1167–1178.
Kamiya A, Tan PL, Kubo K, Engelhard C, Ishizuka K, Kubo A et al (2008). Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch Gen Psychiatry 65: 996–1006.
Kandalepas PC, Sadleir KR, Eimer WA, Zhao J, Nicholson DA, Vassar R (2013). The Alzheimer's beta-secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol 126: 329–352.
Kiyota T, Okuyama S, Swan RJ, Jacobsen MT, Gendelman HE, Ikezu T (2010). CNS expression of anti-inflammatory cytokine interleukin-4 attenuates Alzheimer's disease-like pathogenesis in APP+PS1 bigenic mice. FASEB J 24: 3093–3102.
Koh YH, von Arnim CA, Hyman BT, Tanzi RE, Tesco G (2005). BACE is degraded via the lysosomal pathway. J Biol Chem 280: 32499–32504.
Koike H, Arguello PA, Kvajo M, Karayiorgou M, Gogos JA (2006). Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc Natl Acad Sci USA 103: 3693–3697.
Kvajo M, McKellar H, Arguello PA, Drew LJ, Moore H, MacDermott AB et al (2008). A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci USA 105: 7076–7081.
Kvajo M, McKellar H, Drew LJ, Lepagnol-Bestel AM, Xiao L, Levy RJ et al (2011). Altered axonal targeting and short-term plasticity in the hippocampus of Disc1 mutant mice. Proc Natl Acad Sci USA 108: E1349–E1358.
Lee EB, Zhang B, Liu K, Greenbaum EA, Doms RW, Trojanowski JQ et al (2005). BACE overexpression alters the subcellular processing of APP and inhibits Abeta deposition in vivo. J Cell Biol 168: 291–302.
Lee FH, Fadel MP, Preston-Maher K, Cordes SP, Clapcote SJ, Price DJ et al (2011). Disc1 point mutations in mice affect development of the cerebral cortex. J Neurosci 31: 3197–3206.
Liang B, Duan BY, Zhou XP, Gong JX, Luo ZG (2010). Calpain activation promotes BACE1 expression, amyloid precursor protein processing, and amyloid plaque formation in a transgenic mouse model of Alzheimer disease. J Biol Chem 285: 27737–27744.
Liu Z, Spirek M, Thornton J, Butow RA (2005). A novel degron-mediated degradation of the RTG pathway regulator, Mks1p, by SCFGrr1. Mol Biol Cell 16: 4893–4904.
Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P et al (2001). Mice deficient in BACE1, the Alzheimer's beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci 4: 231–232.
Lyketsos CG, Carrillo MC, Ryan JM, Khachaturian AS, Trzepacz P, Amatniek J et al (2011). Neuropsychiatric symptoms in Alzheimer's disease. Alzheimers Dement 7: 532–539.
Ma TM, Abazyan S, Abazyan B, Nomura J, Yang C, Seshadri S et al (2013). Pathogenic disruption of DISC1-serine racemase binding elicits schizophrenia-like behavior via D-serine depletion. Mol Psychiatry 18: 557–567.
Masters CL, Beyreuther K (2006). Alzheimer's centennial legacy: prospects for rational therapeutic intervention targeting the Abeta amyloid pathway. Brain 129: 2823–2839.
Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T et al (2003). Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry 8: 685–694.
Ohno M, Sametsky EA, Younkin LH, Oakley H, Younkin SG, Citron M et al (2004). BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer's disease. Neuron 41: 27–33.
Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K et al (2003). Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA 100: 289–294.
Park YU, Jeong J, Lee H, Mun JY, Kim JH, Lee JS et al (2010). Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc Natl Acad Sci USA 107: 17785–17790.
Petrus E, Lee HK (2014). BACE1 is necessary for experience-dependent homeostatic synaptic plasticity in visual cortex. Neural Plast 2014: 128631.
Pletnikov MV, Ayhan Y, Nikolskaia O, Xu Y, Ovanesov MV, Huang H et al (2008). Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry 13: 173–186 115.
Plucinska K, Crouch B, Koss D, Robinson L, Siebrecht M, Riedel G et al (2014). Knock-in of human BACE1 cleaves murine APP and reiterates Alzheimer-like phenotypes. J Neurosci 34: 10710–10728.
Reiserer RS, Harrison FE, Syverud DC, McDonald MP (2007). Impaired spatial learning in the APPSwe+PSEN1DeltaE9 bigenic mouse model of Alzheimer's disease. Genes Brain Behav 6: 54–65.
Sapra M, Kim KY (2009). Anti-amyloid treatments in Alzheimer's disease. Recent Pat CNS Drug Discov 4: 143–148.
Sathya M, Premkumar P, Karthick C, Moorthi P, Jayachandran KS, Anusuyadevi M (2012). BACE1 in Alzheimer's disease. Clin Chim Acta 414: 171–178.
Schosser A, Gaysina D, Cohen-Woods S, Chow PC, Martucci L, Craddock N et al (2010). Association of DISC1 and TSNAX genes and affective disorders in the depression case-control (DeCC) and bipolar affective case-control (BACCS) studies. Mol Psychiatry 15: 844–849.
Seshadri S, Kamiya A, Yokota Y, Prikulis I, Kano S, Hayashi-Takagi A et al (2010). Disrupted-in-Schizophrenia-1 expression is regulated by beta-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc Natl Acad Sci USA 107: 5622–5627.
Shahani N, Seshadri S, Jaaro-Peled H, Ishizuka K, Hirota-Tsuyada Y, Wang Q et al (2015). DISC1 regulates trafficking and processing of APP and Abeta generation. Mol Psychiatry 20: 874–879.
Sisodia SS (1992). Beta-amyloid precursor protein cleavage by a membrane-bound protease. Proc Natl Acad Sci USA 89: 6075–6079.
Thinakaran G, Teplow DB, Siman R, Greenberg B, Sisodia SS (1996). Metabolism of the "Swedish" amyloid precursor protein variant in neuro2a (N2a) cells. Evidence that cleavage at the "beta-secretase" site occurs in the golgi apparatus. J Biol Chem 271: 9390–9397.
Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P et al (1999). Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286: 735–741.
Vassar R, Kovacs DM, Yan R, Wong PC (2009). The beta-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J Neurosci 29: 12787–12794.
Wang J, Shan C, Cao W, Zhang C, Teng J, Chen J (2013). SCG10 promotes non-amyloidogenic processing of amyloid precursor protein by facilitating its trafficking to the cell surface. Hum Mol Genet 22: 4888–4900.
Yan R, Han P, Miao H, Greengard P, Xu H (2001). The transmembrane domain of the Alzheimer's beta-secretase (BACE1) determines its late Golgi localization and access to betaamyloid precursor protein (APP) substrate. J Biol Chem 276: 36788–36796.
Yang LB, Lindholm K, Yan R, Citron M, Xia W, Yang XL et al (2003). Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med 9: 3–4.
Yin YI, Bassit B, Zhu L, Yang X, Wang C, Li YM (2007). {gamma}-Secretase Substrate Concentration Modulates the Abeta42/Abeta40 Ratio: IMPLICATIONS FOR ALZHEIMER DISEASE. J Biol Chem 282: 23639–23644.
Young-Pearse TL, Suth S, Luth ES, Sawa A, Selkoe DJ (2010). Biochemical and functional interaction of disrupted-in-schizophrenia 1 and amyloid precursor protein regulates neuronal migration during mammalian cortical development. J Neurosci 30: 10431–10440.
Yu YX, Shen L, Xia P, Tang YW, Bao L, Pei G (2006). Syntaxin 1A promotes the endocytic sorting of EAAC1 leading to inhibition of glutamate transport. J Cell Sci 119: 3776–3787.
Zhang MY, Zheng CY, Zou MM, Zhu JW, Zhang Y, Wang J et al (2014). Lamotrigine attenuates deficits in synaptic plasticity and accumulation of amyloid plaques in APP/PS1 transgenic mice. Neurobiol Aging 35: 2713–2725.
Acknowledgements
This work was supported by grants to QH Ma from the National Program on Key Basic Research Project (2013CB945602), the National Natural Science Foundation of China (31171313 and 81271424), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Soochow University Startup Foundation (Q421500110), to RX Xu from the Millitary Medical Project (BMS11J002). MS is supported by the Shantou University Medical School.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Deng, QS., Dong, XY., Wu, H. et al. Disrupted-in-Schizophrenia-1 Attenuates Amyloid-β Generation and Cognitive Deficits in APP/PS1 Transgenic Mice by Reduction of β-Site APP-Cleaving Enzyme 1 Levels. Neuropsychopharmacol 41, 440–453 (2016). https://doi.org/10.1038/npp.2015.164
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2015.164
This article is cited by
-
The plasma peptides of Alzheimer’s disease
Clinical Proteomics (2021)
-
Functional brain defects in a mouse model of a chromosomal t(1;11) translocation that disrupts DISC1 and confers increased risk of psychiatric illness
Translational Psychiatry (2021)
-
Bis(9)-(−)-Meptazinol, a novel dual-binding AChE inhibitor, rescues cognitive deficits and pathological changes in APP/PS1 transgenic mice
Translational Neurodegeneration (2018)
-
The Mitochondrion: A Potential Therapeutic Target for Alzheimer’s Disease
Neuroscience Bulletin (2018)