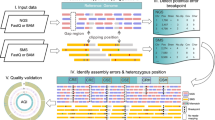
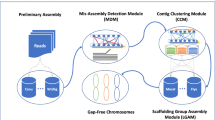
Abstract
Genome projects now produce draft assemblies within weeks owing to advanced high-throughput sequencing technologies. For milestone projects such as Escherichia coli or Homo sapiens, teams of scientists were employed to manually curate and finish these genomes to a high standard. Nowadays, this is not feasible for most projects, and the quality of genomes is generally of a much lower standard. This protocol describes software (PAGIT) that is used to improve the quality of draft genomes. It offers flexible functionality to close gaps in scaffolds, correct base errors in the consensus sequence and exploit reference genomes (if available) in order to improve scaffolding and generating annotations. The protocol is most accessible for bacterial and small eukaryotic genomes (up to 300 Mb), such as pathogenic bacteria, malaria and parasitic worms. Applying PAGIT to an E. coli assembly takes ∼24 h: it doubles the average contig size and annotates over 4,300 gene models.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Chain, P.S. et al. Genome project standards in a new era of sequencing. Science 326, 236–237 (2009).
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Brent, M.R. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat. Rev. Genet. 9, 62–73 (2008).
Pruitt, K.D., Tatusova, T., Brown, G.R. & Maglott, D.R. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 (2012).
Salzberg, S.L. et al. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22, 557–567 (2012).
Narzisi, G. & Mishra, B. Comparing de novo genome assembly: the long and short of it. PLos ONE 6, e19175 (2011).
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008).
Zhang, J., Chiodini, R., Badr, A. & Zhang, G. The impact of next-generation sequencing on genomics. J. Genet. Genomics 38, 95–109 (2011).
Alkan, C., Sajjadian, S. & Eichler, E.E. Limitations of next-generation genome sequence assembly. Nat. Methods 8, 61–65 (2011).
Miller, J.R., Koren, S. & Sutton, G. Assembly algorithms for next-generation sequencing data. Genomics 6, 315–327 (2010).
Treangen, T.J., Sommer, D.D., Angly, F.E., Koren, S. & Pop, M. Next generation sequence assembly with AMOS. Curr. Protoc. Bioinform. 33, 11.8.1–11.8.18 (2011).
Zerbino, D.R. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinform. 31, 11.5.1–11.5.12 (2010).
Assefa, S., Keane, T.M., Otto, T.D., Newbold, C. & Berriman, M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25, 1968–1969 (2009).
Tsai, I.J., Otto, T.D. & Berriman, M. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 11, R41 (2010).
Otto, T.D., Sanders, M., Berriman, M. & Newbold, C. Iterative correction of reference nucleotides (iCORN) using second generation sequencing technology. Bioinformatics 26, 1704–1707 (2010).
Otto, T.D., Dillon, G.P., Degrave, W.S. & Berriman, M. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 39, e57 (2011).
Croucher, N.J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 (2011).
Downing, T. et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 21, 2143–2156 (2011).
Rogers, M.B.H. et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 21, 2129–2142 (2011).
Protasio, A. et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 6, e1455 (2012).
Kikuchi, T. et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7, e1002219 (2011).
Olson, P.D., Zarowiecki, M., Kiss, F. & Brehm, K. Cestode genomics—progress and prospects for advancing basic and applied aspects of flatworm biology. Parasite Immunol. 34, 130–150 (2011).
Heilbronner, S. et al. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 322, 60–67 (2011).
Omer, H. et al. Genotypic and phenotypic modifications of Neisseria meningitidis after an accidental human passage. PLoS One 6, e17145 (2011).
Petty, N.K. et al. Citrobacter rodentium is an unstable pathogen showing evidence of significant genomic flux. PLoS Pathog. 7, e1002018 (2011).
Stabler, R.A. et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10, R102 (2009).
Kurtz, S. et al. Verstile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Carver, T.B. et al. Artemis and ACT viewing, annotation and comparing sequences stored in relational database. Bioinformatics 24, 2672–2676 (2008).
Koressaar, T. & Remm, M. Enhancements and modifications for primer design program Primer3. Bioinformatics 23, 1289–1291 (2007).
Galardini, M., Biondi, G., Bazzicalupo, M. & Mengoni, A. CONTIGuator: a bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol. Med. 6, 11 (2011).
van Hijum, S., Zomer, A., Kuipers, O. & Kok, J. Projector 2: contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acid Res. 33, W560–W566 (2005).
Richter, D., Schuster, S. & Huson, D. OSLay: optimal syntenic layout of unfinished assemblies. Bioinformatics 23, 1573–1579 (2007).
Husemann, P. & Stoye, J. r2cat: synteny plots and comparative assembly. Bioinformatics 26, 570–571 (2010).
Li, R. et al. The sequence and de novo assembly of the giant panda genome. Nature 463, 311–317 (2010).
Yao, G. et al. Graph accordance of next-generation sequence assemblies. Bioinformatics 28, 13–16 (2012).
Zimin, A.V., Smith, D.R., Sutton, G. & Yorke, J.A. Assembly reconciliation. Bioinformatics 24, 42–45 (2008).
Yang, X., Medvin, D., Narasimham, G., Yoder-Himes, D. & Lory, S. CloG: a pipeline for closing gaps in a draft assembly using short reads. in 2011 IEEE 1st International Conference on Computational Advances in Bio and Medical Sciences (Orlando, Florida) 202–207 (IEEE, 2011).
Pop, M., Kosack, D. & Salzberg, S. Hierarchical scaffolding with bambus. Genome Res. 14, 149–159 (2004).
Dayarian, A., Michael, T. & Sengupta, A. SOPRA: scaffolding algorithm for paired reads via statistical optimization. BMC Bioinformatics 11, 345 (2010).
Boetzer, M., Henkel, C., Jansen, H., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011).
Gao, S., Nagarajan, H. & Sung, W. Opera: reconstructing optimal genomic scaffolds using pair-end sequences. Res. Comput. Mol. Biol. 6577, 437–451 (2011).
Ronaghi, M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 11, 3–11 (2001).
Ning, Z., Cox, A. & Mullikin, J. SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1724–1729 (2001).
Manske, H. & Kwiatkowski, D. SNP-o-matic. Bioinformatics 25, 2434–2435 (2009).
Gajer, P.S., Schatz, M. & Salzberg, S.L. Automated correction of genome sequence errors. Nucleic Acids Res. 32, 562–569 (2004).
Dutilh, B.H., Huynen, M.A. & Strous, M. Increasing the coverage of a metapopulation consensus genome by iterative read mapping assembly. Bioinformatics 25, 2878–2881 (2009).
Hubbard, T.J. et al. Ensembl 2009. Nucleic Acid Res. 37, D690–D697 (2009).
Davila, S.M. et al. GARSA: genomic analysis resources for sequence annotation. Bioinformatics 21, 4302–4303 (2005).
Almeida, L. et al. A system for automated bacterial (genome) integrated annotation—SABIA. Bioinformatics 20, 2832–2833 (2004).
Markowitz, V.M. et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38, D382–D390 (2010).
Stanke, M. & Morgenstern, B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 22, W465–W467 (2005).
Thomson, N.R.H. et al. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18, 161–171 (2008).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Phillippy, A., Schatz, M.C. & Pop, M. Genome assembly forensics: finding the elusive mis-assembly. Genome Biol. 9, R55 (2008).
Acknowledgements
We thank L. Chappel for testing and checking the protocol; T. Carver for helping with the installation of the Virtual Machine; and M. Hunt for testing the virtual machine. T.D.O. was supported by the European Union 7th framework European Virtual Institute of Malaria Research (EVIMalaR); I.J.T., S.A.A. and M.B. were supported by the Wellcome Trust (grant number: 098051).
Author information
Authors and Affiliations
Contributions
M.T.S., T.D.O., C.N. and M.B. conceived and executed the examples. T.D.O., M.T.S., I.J.T. and S.A.A. conceived and wrote the installation procedures. All authors were involved with the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
PAGIT: two worked examples. Here we give a synopsis of the work-flow used for the two examples discussed in the "Anticipated Results" section. (DOCX 31 kb)
Rights and permissions
About this article
Cite this article
Swain, M., Tsai, I., Assefa, S. et al. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat Protoc 7, 1260–1284 (2012). https://doi.org/10.1038/nprot.2012.068
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.068
This article is cited by
-
Genomic analysis of Leptospira interrogans serovar Paidjan and Dadas isolates from carrier dogs and comparative genomic analysis to detect genes under positive selection
BMC Genomics (2019)
-
New insights from Opisthorchis felineus genome: update on genomics of the epidemiologically important liver flukes
BMC Genomics (2019)
-
Whole genome sequencing of the monomorphic pathogen Mycobacterium bovis reveals local differentiation of cattle clinical isolates
BMC Genomics (2018)
-
Staphylococcus aureus from patients with chronic rhinosinusitis show minimal genetic association between polyp and non-polyp phenotypes
BMC Ear, Nose and Throat Disorders (2018)
-
The origin and adaptive evolution of domesticated populations of yeast from Far East Asia
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.