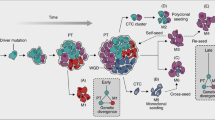
Abstract
Recently, microRNAs (miRNAs) have been discovered to have a role in metastasis. Here we describe how miRNAs are involved in advanced stages of tumour progression, stressing their roles as metastasis activators or suppressors, and discuss their possible use in the clinic as predictive markers and as therapeutic strategies for patients with metastases. Furthermore, we develop the concept that the same miRNAs could be involved both in the cancer stem cell phenotype and in the ability of specific cancer cells to produce metastases, thus representing a mechanistic link between the initial and the final steps of tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Eccles, S. A. & Welch, D. R. Metastasis: recent discoveries and novel treatment strategies. Lancet 369, 1742–1757 (2007).
Nguyen, D. X. & Massague, J. Genetic determinants of cancer metastasis. Nature Rev. Genet. 8, 341–352 (2007).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nature Rev. Cancer 6, 857–866 (2006).
Calin, G. A. & Croce, C. M. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 66, 7390–7394 (2006).
Esquela-Kerscher, A. & Slack, F. J. Oncomirs — microRNAs with a role in cancer. Nature Rev. Cancer 6, 259–269 (2006).
Hammond, S. M. MicroRNAs as tumor suppressors. Nature Genet. 39, 582–583 (2007).
Chivukula, R. R. & Mendell, J. T. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem. Sci. 33, 474–481 (2008).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Lujambio, A. & Esteller, M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle 6, 1455–1459 (2007).
Calin, G. A. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA 101, 2999–3004 (2004).
Chang, T. C. et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752 (2007).
Chang, T. C. et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genet. 40, 43–50 (2008).
Lodygin, D. et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7, 2591–2600 (2008).
Wei, J. S. et al. The MYCN oncogene is a direct target of miR-34a. Oncogene 27, 5204–5213 (2008).
He, L., He, X., Lowe, S. W. & Hannon, G. J. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nature Rev. Cancer 7, 819–822 (2007).
O'Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–43 (2005).
Lotterman, C. D., Kent, O. A. & Mendell, J. T. Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle 7, 2493–2499 (2008).
Ma, L., Teruya-Feldstein, J. & Weinberg, R. A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682–688 (2007).
Kong, W. et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 28, 6773–6784 (2008).
Lujambio, A. et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA 105, 13556–13561 (2008).
Iorio, M. V. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070 (2005).
Yang, J., Mani, S. A. & Weinberg, R. A. Exploring a new twist on tumor metastasis. Cancer Res. 66, 4549–4552 (2006).
Gee, H. E. et al. MicroRNA-10b and breast cancer metastasis. Nature 455, E8–E9; author reply E9 (2008).
Myers, C., Charboneau, A., Cheung, I., Hanks, D. & Boudreau, N. Sustained expression of homeobox D10 inhibits angiogenesis. Am. J. Pathol. 161, 2099–2109 (2002).
Voorhoeve, P. M. et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181 (2006).
Huang, Q. et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature Cell Biol. 10, 202–210 (2008).
Zhu, S. et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18, 350–359 (2008).
Volinia, S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA 103, 2257–2261 (2006).
Zhu, S., Si, M. L., Wu, H. & Mo, Y. Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 282, 14328–14336 (2007).
Asangani, I. A. et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136 (2008).
Gabriely, G. et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 28, 5369–5380 (2008).
Meng, F. et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 (2007).
Tavazoie, S. F. et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 (2008).
Hoser, M. et al. Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J. Neurosci. 27, 5495–5505 (2007).
Liao, Y. L. et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene 27, 5578–5589 (2008).
Orend, G. & Chiquet-Ehrismann, R. Tenascin-C induced signaling in cancer. Cancer Lett. 244, 143–163 (2006).
Crawford, M. et al. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem. Biophys. Res. Commun. 373, 607–612 (2008).
Feller, S. M. Crk family adaptors-signalling complex formation and biological roles. Oncogene 20, 6348–6371 (2001).
Joyce, J. A. & Pollard, J. W. Microenvironmental regulation of metastasis. Nature Rev. Cancer (in the press).
Sengupta, S. et al. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc. Natl Acad. Sci. USA 105, 5874–5878 (2008).
Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. A molecular signature of metastasis in primary solid tumors. Nature Genet. 33, 49–54 (2003).
Friedl, P. & Wolf, K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nature Rev. Cancer 3, 362–374 (2003).
Sahai, E. & Marshall, C. J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nature Cell Biol. 5, 711–719 (2003).
Lin, S. L., Chiang, A., Chang, D. & Ying, S. Y. Loss of mir-146a function in hormone-refractory prostate cancer. RNA 14, 417–424 (2008).
Bhaumik, D. et al. Expression of microRNA-146 suppresses NF-κB activity with reduction of metastatic potential in breast cancer cells. Oncogene 27, 5643–5647 (2008).
Naugler, W. E. & Karin, M. NF-κB and cancer-identifying targets and mechanisms. Curr. Opin. Genet. Dev. 18, 19–26 (2008).
Rodriguez, P. L., Sahay, S., Olabisi, O. O. & Whitehead, I. P. ROCK I-mediated activation of NF-κB by RhoB. Cell Signal. 19, 2361–2369 (2007).
Visvader, J. E. & Lindeman, G. J. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nature Rev. Cancer 8, 755–768 (2008).
Hatfield, S. & Ruohola-Baker, H. microRNA and stem cell function. Cell Tissue Res. 331, 57–66 (2008).
Calabrese, J. M., Seila, A. C., Yeo, G. W. & Sharp, P. A. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 104, 18097–18102 (2007).
Marson, A. et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533 (2008).
Polyak, K. & Weinberg, R. A. Epithelial and mesenchymal transitions: acquisition of malignant and stem cell traits. Nature Rev. Cancer 5 Mar 2009 (doi: 10.1038/nrc2620).
Mani, S. A. et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008).
Thiery, J. P. Epithelial–mesenchymal transitions in tumour progression. Nature Rev. Cancer 2, 442–454 (2002).
Friedl, P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr. Opin. Cell Biol. 16, 14–23 (2004).
Ansieau, S. et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 14, 79–89 (2008).
Dalerba, P. & Clarke, M. F. Cancer stem cells and tumor metastasis: first steps into uncharted territory. Cell Stem Cell 1, 241–242 (2007).
Costinean, S. et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc. Natl Acad. Sci. USA 103, 7024–7029 (2006).
Yu, F. et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 (2007).
Greco, S. J. & Rameshwar, P. MicroRNAs regulate synthesis of the neurotransmitter substance P in human mesenchymal stem cell-derived neuronal cells. Proc. Natl Acad. Sci. USA 104, 15484–15489 (2007).
Varambally, S. et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699 (2008).
Budhu, A. et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology 47, 897–907 (2008).
Fletcher, A. M., Heaford, A. C. & Trask, D. K. Detection of metastatic head and neck squamous cell carcinoma using the relative expression of tissue-specific mir-205. Transl. Oncol. 1, 202–208 (2008).
Nass, D. et al. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can. be used to differentiate primary from metastatic brain tumors. Brain Pathol. 8 Jul 2008 (doi:10.1111/j.1750-3639.2008.00184.x).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Rosenfeld, N. et al. MicroRNAs accurately identify cancer tissue origin. Nature Biotechnol. 26, 462–469 (2008).
Tili, E. et al. miRNAs and their potential for use against cancer and other diseases. Future Oncol. 3, 521–537 (2007).
Calin, G. A. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229 (2007).
Filipowicz, W., Bhattacharyya, S. N. & Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev. Genet. 9, 102–114 (2008).
Valencia-Sanchez, M. A., Liu, J., Hannon, G. J. & Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524 (2006).
Vasudevan, S., Tong, Y. & Steitz, J. A. Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934 (2007).
Orom, U. A., Nielsen, F. C. & Lund, A. H. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 (2008).
Tay, Y., Zhang, J., Thomson, A. M., Lim, B. & Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–1128 (2008).
Hwang, H. W., Wentzel, E. A. & Mendell, J. T. A hexanucleotide element directs microRNA nuclear import. Science 315, 97–100 (2007).
Place, R. F., Li, L. C., Pookot, D., Noonan, E. J. & Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA 105, 1608–1613 (2008).
Gregory, P. A., Bracken, C. P., Bert, A. G. & Goodall, G. J. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 7, 3112–3118 (2008).
Peinado, H., Olmeda, D. & Cano, A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature Rev. Cancer 7, 415–428 (2007).
Burk, U. et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 (2008).
Gregory, P. A. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biol. 10, 593–601 (2008).
Park, S. M., Gaur, A. B., Lengyel, E. & Peter, M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 (2008).
Korpal, M., Lee, E. S., Hu, G. & Kang, Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 (2008).
Bracken, C. P. et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition. Cancer Res. 68, 7846–7854 (2008).
Yan, L. X. et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14, 2348–2360 (2008).
Gleave, M. E. & Monia, B. P. Antisense therapy for cancer. Nature Rev. Cancer. 5, 468–479 (2005).
Opalinska, J. B. & Gewirtz, A. M. Nucleic-acid therapeutics: basic principles and recent applications. Nature Rev. Drug Discov. 1, 503–514 (2002).
Esau, C. C. Inhibition of microRNA with antisense oligonucleotides. Methods 44, 55–60 (2008).
Naguibneva, I. et al. An LNA-based loss-of-function assay for micro-RNAs. Biomed. Pharmacother. 60, 633–638 (2006).
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 438, 685–689 (2005).
Kulshreshtha, R. et al. A microRNA signature of hypoxia. Mol. Cell. Biol. 27, 1859–1867 (2007).
Mertens-Talcott, S. U., Chintharlapalli, S., Li, X. & Safe, S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcriptionfactors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 67, 11001–11011 (2007).
Pulkkinen, K., Malm, T., Turunen, M., Koistinaho, J. & Yla-Herttuala, S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 582, 2397–2401 (2008).
Hua, Z. et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE 1, e116 (2006).
le Sage, C. et al. Regulation of the p27Kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 26, 3699–3708 (2007).
Baldassarre, G. et al. p27Kip1–stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell 7, 51–63 (2005).
Fish, J. E. et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 15, 272–284 (2008).
Dews, M. et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nature Genet. 38, 1060–1065 (2006).
Kuehbacher, A., Urbich, C., Zeiher, A. M. & Dimmeler, S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 101, 59–68 (2007).
Urbich, C., Kuehbacher, A. & Dimmeler, S. Role of microRNAs in vascular diseases, inflammation and angiogenesis. Cardiovasc. Res. 79, 581–588 (2008).
Roldo, C. et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 24, 4677–4684 (2006).
Loffler, D. et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 110, 1330–1333 (2007).
Wang, S. et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271 (2008).
Harris, T. A., Yamakuchi, M., Ferlito, M., Mendell, J. T. & Lowenstein, C. J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl Acad. Sci. USA 105, 1516–1521 (2008).
Saito, Y. et al. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem. Biophys. Res. Commun. 379, 726–731 (2009).
Taganov, K. D., Boldin, M. P., Chang, K. J. & Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA 103, 12481–12486 (2006).
Hurst, D. R. et al. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 69, 1279–1283 (2009).
Pekarsky, Y. et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res 66, 11590–11593 (2006).
Yanaihara, N. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 (2006).
Mayr, C., Hemann, M. T. & Bartel, D. P. Disrupting the pairing between let-7 and Hmga2 enhances oncogene transformation. Science 315, 1576–1579 (2007).
Acknowledgements
G.A.C. is supported as a Fellow at The University of Texas M. D. Anderson Research Trust, and as a Fellow of The University of Texas System Regents Research Scholar and by the Ladjevardian Regents Research Scholar Fund. This study was supported in part by an Institutional Research Grant, by a Cancer Center Support Grant (New Faculty Award), by a CTT/3I-TD grant and by a Breast Cancer SPORE Developmental Award to G.A.C.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
miRBase
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Nicoloso, M., Spizzo, R., Shimizu, M. et al. MicroRNAs — the micro steering wheel of tumour metastases. Nat Rev Cancer 9, 293–302 (2009). https://doi.org/10.1038/nrc2619
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc2619
This article is cited by
-
A novel therapeutic strategy: the significance of exosomal miRNAs in acute myeloid leukemia
Medical Oncology (2024)
-
LncRNA34977 promotes the proliferation, migration, and invasion and inhibits the apoptosis of canine mammary tumors by regulating the expression of miR-8881/ELAVL4
Functional & Integrative Genomics (2023)
-
The relationship between microRNAs and bladder cancer: are microRNAs useful to predict bladder cancer in suspicious patients?
International Urology and Nephrology (2023)
-
MiR-124-3p inhibits tumor progression in prostate cancer by targeting EZH2
Functional & Integrative Genomics (2023)
-
p63, a key regulator of Ago2, links to the microRNA-144 cluster
Cell Death & Disease (2022)