Key Points
-
RecQ helicases are a highly conserved family and are considered to be genome 'caretakers' that maintain chromosome stability and suppress neoplastic transformation.
-
The crystal structure of RecQ helicases reveals several conserved functional domains. Some of these domains define a particular member of the RecQ helicase family and may be important for functionally distinguishing between the different RecQ helicases expressed in a particular organism.
-
Defects in three of the five human RecQ helicase members give rise to at least five defined disorders associated with cancer predisposition, premature ageing and developmental abnormalities. Mutations in BLM and WRN lead to Bloom's and Werner's syndromes, respectively, and RECQ4 is associated with three distinct disorders: Rothmund–Thomson, RAPADILINO and Baller–Gerold syndromes. These RECQ4 disorders display the common feature of an abnormality in bone development.
-
Mouse models of human RecQ helicase disorders have been generated that partially recapitulate the phenotypes seen in human patients.
-
RecQ helicases possess several biochemical activities and have several important roles in DNA replication and recombination. Some of these activities require or are modulated by physical interactions with other nuclear proteins.
-
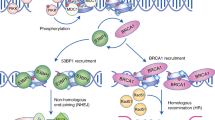
BLM is proposed to have roles in mitosis that resolve late-replication intermediates in conjunction with topoisomerase IIIα. Together, these proteins probably function to decatenate entangled DNA that arises during DNA replication.
Abstract
Around 1% of the open reading frames in the human genome encode predicted DNA and RNA helicases. One highly conserved group of DNA helicases is the RecQ family. Genetic defects in three of the five human RecQ helicases, BLM, WRN and RECQ4, give rise to defined syndromes associated with cancer predisposition, some features of premature ageing and chromosomal instability. In recent years, there has been a tremendous advance in our understanding of the cellular functions of individual RecQ helicases. In this Review, we discuss how these proteins might suppress genomic rearrangements, and therefore function as 'caretaker' tumour suppressors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Soultanas, P. & Wigley, D. B. Unwinding the 'Gordian knot' of helicase action. Trends Biochem. Sci. 26, 47–54 (2001).
Wu, L. & Hickson, I. D. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40, 279–306 (2006).
Hanada, K. & Hickson, I. D. Molecular genetics of RecQ helicase disorders. Cell. Mol. Life Sci. 64, 2306–2322 (2007).
Bachrati, C. Z. & Hickson, I. D. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 117, 219–233 (2008).
Hanada, K. et al. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl Acad. Sci. USA 94, 3860–3865 (1997).
Sharma, S., Doherty, K. M. & Brosh, R. M. Jr. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 398, 319–337 (2006).
Hartung, F. & Puchta, H. The RecQ gene family in plants. J. Plant Physiol. 163, 287–296 (2006).
Shimamoto, A., Nishikawa, K., Kitao, S. & Furuichi, Y. Human RecQ5β, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3α and 3β. Nucleic Acids Res. 28, 1647–1655 (2000).
Guo, R. B., Rigolet, P., Zargarian, L., Fermandjian, S. & Xi, X. G. Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom's syndrome helicase. Nucleic Acids Res. 33, 3109–3124 (2005).
Bennett, R. J. & Keck, J. L. Structure and function of RecQ DNA helicases. Crit. Rev. Biochem. Mol. Biol. 39, 79–97 (2004).
Lee, J. W., Harrigan, J., Opresko, P. L. & Bohr, V. A. Pathways and functions of the Werner syndrome protein. Mech. Ageing Dev. 126, 79–86 (2005).
Morozov, V., Mushegian, A. R., Koonin, E. V. & Bork, P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem. Sci. 22, 417–418 (1997).
Bernstein, D. A. & Keck, J. L. Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family. Nucleic Acids Res. 31, 2778–2785 (2003).
Rodriguez, A. C. & Stock, D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 21, 418–426 (2002).
Lima, C. D., Wang, J. C. & Mondragon, A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature 367, 138–146 (1994).
Bernstein, D. A., Zittel, M. C. & Keck, J. L. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 22, 4910–4921 (2003).
Bernstein, D. A. & Keck, J. L. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure 13, 1173–1182 (2005).
Opresko, P. L. Telomere ResQue and preservation — roles for the Werner syndrome protein and other RecQ helicases. Mech. Ageing Dev. 129, 79–90 (2008).
Brosh, R. M. Jr & Bohr, V. A. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 35, 7527–7544 (2007).
Zhu, J. et al. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 283, 29405–29415 (2008).
Eladad, S. et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 14, 1351–1365 (2005).
Wu, L., Davies, S. L., Levitt, N. C. & Hickson, I. D. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276, 19375–19381 (2001).
Kawabe, Y. et al. Covalent modification of the Werner's syndrome gene product with the ubiquitin-related protein, SUMO-1. J. Biol. Chem. 275, 20963–20966 (2000).
Rog, O., Miller, K. M., Ferreira, M. G. & Cooper, J. P. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol. Cell 33, 559–569 (2009).
Hickson, I. D. RecQ helicases: caretakers of the genome. Nature Rev. Cancer 3, 169–178 (2003).
Levitt, N. C. & Hickson, I. D. Caretaker tumour suppressor genes that defend genome integrity. Trends Mol. Med. 8, 179–186 (2002).
Grandori, C. et al. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 17, 1569–1574 (2003).
Ellis, N. A. et al. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83, 655–666 (1995).
Yu, C. E. et al. Positional cloning of the Werner's syndrome gene. Science 272, 258–262 (1996).
Kitao, S. et al. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nature Genet. 22, 82–84 (1999).
Siitonen, H. A. et al. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 12, 2837–2844 (2003).
Van Maldergem, L. et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 43, 148–152 (2006). References 30–32 showed that RECQ4 is mutated in three distinct disorders: Rothmund-Thomson syndrome, RAPADILINO and Baller-Gerold syndrome.
German, J., Sanz, M. M., Ciocci, S., Ye, T. Z. & Ellis, N. A. Syndrome-causing mutations of the BLM gene in persons in the Bloom's Syndrome Registry. Hum. Mutat. 28, 743–753 (2007).
Li, L., Eng, C., Desnick, R. J., German, J. & Ellis, N. A. Carrier frequency of the Bloom syndrome blm Ash mutation in the Ashkenazi Jewish population. Mol. Genet. Metab. 64, 286–290 (1998).
Ellis, N. A. et al. The Ashkenazic Jewish Bloom syndrome mutation blmAsh is present in non-Jewish Americans of Spanish ancestry. Am. J. Hum. Genet. 63, 1685–1693 (1998).
German, J., Archibald, R. & Bloom, D. Chromosomal breakage in a rare and probably genetically determined syndrome of Man. Science 148, 506–507 (1965).
Chaganti, R. S., Schonberg, S. & German, J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl Acad. Sci. USA 71, 4508–4512 (1974).
Chester, N., Kuo, F., Kozak, C., O'Hara, C. D. & Leder, P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes Dev. 12, 3382–3393 (1998).
Luo, G. et al. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nature Genet. 26, 424–429 (2000). References 38 and 39 showed that Blm is an essential gene in mice but that mice carrying a hypomorphic allele display cancer predisposition.
McDaniel, L. D. et al. Chromosome instability and tumor predisposition inversely correlate with BLM protein levels. DNA Repair (Amst.) 2, 1387–1404 (2003).
Goss, K. H. et al. Enhanced tumor formation in mice heterozygous for Blm mutation. Science 297, 2051–2053 (2002).
Goto, M., Miller, R. W., Ishikawa, Y. & Sugano, H. Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol. Biomarkers Prev. 5, 239–246 (1996).
Yu, C. E. et al. Mutations in the consensus helicase domains of the Werner syndrome gene. Werner's Syndrome Collaborative Group. Am. J. Hum. Genet. 60, 330–341 (1997).
Salk, D., Bryant, E., Au, K., Hoehn, H. & Martin, G. M. Systematic growth studies, cocultivation, and cell hybridization studies of Werner syndrome cultured skin fibroblasts. Hum. Genet. 58, 310–316 (1981).
Schulz, V. P. et al. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum. Genet. 97, 750–754 (1996).
Lebel, M. & Leder, P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc. Natl Acad. Sci. USA 95, 13097–13102 (1998).
Massip, L. et al. Increased insulin, triglycerides, reactive oxygen species, and cardiac fibrosis in mice with a mutation in the helicase domain of the Werner syndrome gene homologue. Exp. Gerontol. 41, 157–168 (2006).
Lebel, M., Cardiff, R. D. & Leder, P. Tumorigenic effect of nonfunctional p53 or p21 in mice mutant in the Werner syndrome helicase. Cancer Res. 61, 1816–1819 (2001).
Lombard, D. B. et al. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol. Cell. Biol. 20, 3286–3291 (2000).
Moore, G., Knoblaugh, S., Gollahon, K., Rabinovitch, P. & Ladiges, W. Hyperinsulinemia and insulin resistance in Wrn null mice fed a diabetogenic diet. Mech. Ageing Dev. 129, 201–206 (2008).
Chang, S. et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nature Genet. 36, 877–882 (2004). This paper defined telomere maintenance as a key role for the WRN protein.
Siitonen, H. A. et al. The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet. 17, 151–158 (2009).
Der Kaloustian, V. M., McGill, J. J., Vekemans, M. & Kopelman, H. R. Clonal lines of aneuploid cells in Rothmund-Thomson syndrome. Am. J. Med. Genet. 37, 336–339 (1990).
Ying, K. L., Oizumi, J. & Curry, C. J. Rothmund-Thomson syndrome associated with trisomy 8 mosaicism. J. Med. Genet. 27, 258–260 (1990).
Orstavik, K. H., McFadden, N., Hagelsteen, J., Ormerod, E. & van der Hagen, C. B. Instability of lymphocyte chromosomes in a girl with Rothmund-Thomson syndrome. J. Med. Genet. 31, 570–572 (1994).
Lindor, N. M. et al. Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am. J. Med. Genet. 90, 223–228 (2000).
Ichikawa, K., Noda, T. & Furuichi, Y. Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases. Nippon Yakurigaku Zasshi 119, 219–226 (2002).
Hoki, Y. et al. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum. Mol. Genet. 12, 2293–2299 (2003).
Mann, M. B. et al. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 14, 813–825 (2005). References 58 and 59 describe mouse models of Rothmund-Thomson syndrome that reiterate some of the clinical features seen in humans.
Mohaghegh, P., Karow, J. K., Brosh, R. M. Jr, Bohr, V. A. & Hickson, I. D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29, 2843–2849 (2001).
Sharma, S. et al. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 280, 28072–28084 (2005).
Umezu, K., Nakayama, K. & Nakayama, H. Escherichia coli RecQ protein is a DNA helicase. Proc. Natl Acad. Sci. USA 87, 5363–5367 (1990).
Bennett, R. J., Sharp, J. A. & Wang, J. C. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem. 273, 9644–9650 (1998).
Bachrati, C. Z., Borts, R. H. & Hickson, I. D. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 34, 2269–2279 (2006).
Sun, H., Karow, J. K., Hickson, I. D. & Maizels, N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273, 27587–27592 (1998).
Orren, D. K., Theodore, S. & Machwe, A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry 41, 13483–13488 (2002).
Brosh, R. M., Jr. et al. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 275, 23500–23508 (2000).
Popuri, V. et al. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 283, 17766–17776 (2008).
Ozsoy, A. Z., Ragonese, H. M. & Matson, S. W. Analysis of helicase activity and substrate specificity of Drosophila RECQ5. Nucleic Acids Res. 31, 1554–1564 (2003).
Harmon, F. G. & Kowalczykowski, S. C. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 12, 1134–1144 (1998).
Bachrati, C. Z. & Hickson, I. D. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 374, 577–606 (2003).
Cheok, C. F., Wu, L., Garcia, P. L., Janscak, P. & Hickson, I. D. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 33, 3932–3941 (2005).
Meetei, A. R. et al. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23, 3417–3426 (2003). This biochemical study identified several BLM interacting factors and paved the way for the identification of several new genome stability genes.
Machwe, A., Xiao, L., Groden, J., Matson, S. W. & Orren, D. K. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 280, 23397–23407 (2005).
Xu, X. & Liu, Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 28, 568–577 (2009).
Garcia, P. L., Liu, Y., Jiricny, J., West, S. C. & Janscak, P. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 23, 2882–2891 (2004).
Macris, M. A., Krejci, L., Bussen, W., Shimamoto, A. & Sung, P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst.) 5, 172–180 (2006). Along with Reference 75, this study characterized the biochemical properties of RECQ4. The partially contradictory findings in these papers require further clarification.
LeRoy, G., Carroll, R., Kyin, S., Seki, M. & Cole, M. D. Identification of RecQL1 as a Holliday junction processing enzyme in human cell lines. Nucleic Acids Res. 33, 6251–6257 (2005).
Karow, J. K., Constantinou, A., Li, J. L., West, S. C. & Hickson, I. D. The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc. Natl Acad. Sci. USA 97, 6504–6508 (2000).
Constantinou, A. et al. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1, 80–84 (2000).
Yin, J. et al. BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J. 24, 1465–1476 (2005).
Chang, M. et al. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 24, 2024–2033 (2005).
Mullen, J. R., Nallaseth, F. S., Lan, Y. Q., Slagle, C. E. & Brill, S. J. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol. Cell. Biol. 25, 4476–4487 (2005).
Xu, D. et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 22, 2843–2855 (2008).
Singh, T. R. et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 22, 2856–2868 (2008). References 84 and 85 describe the identification of RMI2, a new subunit of the BLM complex required for complex stability.
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J. & Stahl, F. W. The double-strand-break repair model for recombination. Cell 33, 25–35 (1983).
Mimitou, E. P. & Symington, L. S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008).
Zhu, Z., Chung, W. H., Shim, E. Y., Lee, S. E. & Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008).
Nimonkar, A. V., Ozsoy, A. Z., Genschel, J., Modrich, P. & Kowalczykowski, S. C. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl Acad. Sci. USA 105, 16906–16911 (2008). References 87–89 revealed a role for RecQ helicases in promoting the early resection step of DSB repair.
Jinks-Robertson, S., Michelitch, M. & Ramcharan, S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 3937–3950 (1993).
Gangloff, S., McDonald, J. P., Bendixen, C., Arthur, L. & Rothstein, R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14, 8391–8398 (1994).
Watt, P. M., Hickson, I. D., Borts, R. H. & Louis, E. J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144, 935–945 (1996).
Ira, G., Malkova, A., Liberi, G., Foiani, M. & Haber, J. E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115, 401–411 (2003).
Prado, F. & Aguilera, A. Control of cross-over by single-strand DNA resection. Trends Genet. 19, 428–431 (2003).
Inbar, O., Liefshitz, B., Bitan, G. & Kupiec, M. The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 275, 30833–30838 (2000).
Symington, L. S., Kang, L. E. & Moreau, S. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 28, 4649–4656 (2000).
Bugreev, D. V., Yu, X., Egelman, E. H. & Mazin, A. V. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 21, 3085–3094 (2007).
Hu, Y. et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21, 3073–3084 (2007). References 97 and 98 indicate that RecQ helicases might suppress homologous recombination by dismantling RAD51 filaments on DNA.
Wu, L. & Hickson, I. D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003). This paper identified the process of Holliday junction dissolution whereby BLM and TOPOIIIα can suppress crossing over during HR and hence diminish SCEs.
Wu, L. et al. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 24, 2679–2687 (2005).
Raynard, S. et al. Functional role of BLAP75 in BLM-topoisomerase IIIα-dependent Holliday junction processing. J. Biol. Chem. 283, 15701–15708 (2008).
Wu, L. et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl Acad. Sci. USA 103, 4068–4073 (2006).
Raynard, S., Bussen, W. & Sung, P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J. Biol. Chem. 281, 13861–13864 (2006). References 101–103 show that RMI1 is important for Holliday junction dissolution.
Plank, J. L., Wu, J. & Hsieh, T. S. Topoisomerase IIIα and Bloom's helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc. Natl Acad. Sci. USA 103, 11118–11123 (2006).
Oh, S. D. et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272 (2007).
Oh, S. D., Lao, J. P., Taylor, A. F., Smith, G. R. & Hunter, N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell 31, 324–336 (2008).
Jessop, L. & Lichten, M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell 31, 313–323 (2008).
Ge, X. Q., Jackson, D. A. & Blow, J. J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 21, 3331–3341 (2007).
Davies, S. L., North, P. S. & Hickson, I. D. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nature Struct. Mol. Biol. 14, 677–679 (2007).
Davies, S. L., North, P. S., Dart, A., Lakin, N. D. & Hickson, I. D. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol. Cell. Biol. 24, 1279–1291 (2004).
Ralf, C., Hickson, I. D. & Wu, L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281, 22839–22846 (2006).
Machwe, A., Xiao, L., Groden, J. & Orren, D. K. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45, 13939–13946 (2006).
Boddy, M. N. et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537–548 (2001).
Ip, S. C. et al. Identification of Holliday junction resolvases from humans and yeast. Nature 456, 357–361 (2008). Identified the yeast Yen1 and human GEN1 proteins as Holliday junction resolvases.
Heller, R. C. & Marians, K. J. Replisome assembly and the direct restart of stalled replication forks. Nature Rev. Mol. Cell Biol. 7, 932–943 (2006).
Wu, L. Role of the BLM helicase in replication fork management. DNA Repair (Amst.) 6, 936–944 (2007).
Crabbe, L., Verdun, R. E., Haggblom, C. I. & Karlseder, J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306, 1951–1953 (2004).
Huang, P. et al. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11, 125–129 (2001).
Cohen, H. & Sinclair, D. A. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl Acad. Sci. USA 98, 3174–3179 (2001).
Johnson, F. B. et al. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20, 905–913 (2001).
Opresko, P. L. et al. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell 14, 763–774 (2004).
Sangrithi, M. N. et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund–Thomson syndrome. Cell 121, 887–898 (2005). This paper showed that RECQ4 has a domain resembling yeast Sld2 protein, and that it has a direct role in DNA replication.
Masumoto, H., Muramatsu, S., Kamimura, Y. & Araki, H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415, 651–655 (2002).
Rosin, M. P. & German, J. Evidence for chromosome instability in vivo in Bloom syndrome: increased numbers of micronuclei in exfoliated cells. Hum. Genet. 71, 187–191 (1985).
Chan, K. L., North, P. S. & Hickson, I. D. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26, 3397–3409 (2007).
Baumann, C., Korner, R., Hofmann, K. & Nigg, E. A. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114 (2007). Together with reference 125, this study identified ultra-fine anaphase bridges, which had previously escaped detection but are common in dividing human cells.
Wang, L. H., Schwarzbraun, T., Speicher, M. R. & Nigg, E. A. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma 117, 123–135 (2008).
Chan, K. L., Palmai-Pallag, T., Ying, S. & Hickson, I. D. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature Cell Biol. 11, 753–760 (2009). This study revealed that fragile sites are prone to being interlinked in mitosis by DNA bridges.
Sutherland, G. R. Heritable fragile sites on human chromosomes I. Factors affecting expression in lymphocyte culture. Am. J. Hum. Genet. 31, 125–135 (1979).
Durkin, S. G. & Glover, T. W. Chromosome fragile sites. Annu. Rev. Genet. 41, 169–192 (2007).
Suski, C. & Marians, K. J. Resolution of converging replication forks by RecQ and topoisomerase III. Mol. Cell 30, 779–789 (2008). These authors revealed that the combination of RecQ and TOPOIIIα could resolve late-stage DNA replication structures.
Mankouri, H. W. & Hickson, I. D. The RecQ helicase–topoisomerase III–Rmi1 complex: a DNA structure-specific 'dissolvasome'? Trends Biochem. Sci. 32, 538–546 (2007).
Acknowledgements
We thank J. Keck for the RecQ protein structure, members of the Hickson laboratory for helpful discussions, and H. Mankouri, C. Bachrati and P. McHugh for helpful comments on the manuscript. Work in the authors' laboratory is funded by Cancer Research UK and the Bloom's Syndrome Foundation. W.K.C was supported by the Anita Mui 'True Heart' Charity Foundation, the K. P. Tin Foundation Limited and the China Oxford Scholarship Fund.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
OMIM
FURTHER INFORMATION
Glossary
- Globular fold
-
'Globe-like' region of a protein that is normally soluble in aqueous solution.
- Promyelocytic leukaemia nuclear bodies
-
Focal nuclear structures that are defined by the presence of PML, which forms an oncogenic fusion protein in cases of acute promyelocytic leukaemia.
- Nonsense-mediated mRNA decay
-
A quality-control mechanism that selectively degrades mRNAs harbouring premature termination codons.
- Hypomorphic
-
An allele that results in a reduction, but not the elimination, of wild-type levels of gene product or activity. This often causes a less severe phenotype than a loss-of-function (or null) allele.
- Osteoporosis
-
Reduction of bone mineral density.
- Craniosynostosis
-
Abnormal skull and brain growth owing to premature fusion of the cranial sutures.
- Poikiloderma
-
Reticulated (lattice-like) cutaneous plaques.
- Branch migration
-
Movement of a DNA crossover junction, causing symmetrical exchange of DNA strands and symmetrical formation of a DNA heteroduplex.
- OB-fold
-
Oligonucleotide and oligosaccharide-binding fold domain of a protein.
Rights and permissions
About this article
Cite this article
Chu, W., Hickson, I. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer 9, 644–654 (2009). https://doi.org/10.1038/nrc2682
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc2682
This article is cited by
-
PARP1 negatively regulates transcription of BLM through its interaction with HSP90AB1 in prostate cancer
Journal of Translational Medicine (2023)
-
Dynamic alternative DNA structures in biology and disease
Nature Reviews Genetics (2023)
-
Leveraging the replication stress response to optimize cancer therapy
Nature Reviews Cancer (2023)
-
Synchronous lung and multiple soft tissue metastases developed from osteosarcoma of tibia: a rare case report and genetic profile analysis
BMC Musculoskeletal Disorders (2022)
-
RNA-sequencing data-driven dissection of human plasma cell differentiation reveals new potential transcription regulators
Leukemia (2021)