Key Points
-
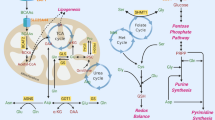
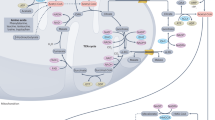
One-carbon metabolism integrates cellular nutrient status by cycling carbon units from amino acid inputs to generate diverse outputs, including redox maintenance and cellular biosynthesis.
-
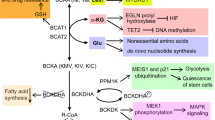
The epigenetic status of cells also seems to be directly linked to one-carbon metabolism through protein and nucleic acid methylation.
-
One-carbon metabolism has long been the focus of antimetabolite-based chemotherapy that includes the agents methotrexate and 5-fluorouracil — two of the most widely used chemotherapies. Additional therapies are currently being explored.
-
Recent findings have provided genetic and functional evidence that multiple nodes in the pathway contain candidate driver genes for oncogenesis.
-
Additional research in one-carbon metabolism may provide biomarkers that would enable advances in patient selection for antimetabolite chemotherapy.
Abstract
One-carbon metabolism involving the folate and methionine cycles integrates nutritional status from amino acids, glucose and vitamins, and generates diverse outputs, such as the biosynthesis of lipids, nucleotides and proteins, the maintenance of redox status and the substrates for methylation reactions. Long considered a 'housekeeping' process, this pathway has recently been shown to have additional complexity. Genetic and functional evidence suggests that hyperactivation of this pathway is a driver of oncogenesis and establishes a link to cellular epigenetic status. Given the wealth of clinically available agents that target one-carbon metabolism, these new findings could present opportunities for translation into precision cancer medicine.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
DeBerardinis, R. J., Lum, J. J., Hatzivassiliou, G. & Thompson, C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell. Metab. 7, 11–20 (2008).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Locasale, J. W. & Cantley, L. C. Metabolic flux and the regulation of mammalian cell growth. Cell. Metab. 14, 443–451 (2011).
Cairns, R. A., Harris, I. S. & Mak, T. W. Regulation of cancer cell metabolism. Nature Rev. Cancer 11, 85–95 (2011).
Ward, P. S. & Thompson, C. B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308 (2012).
Schulze, A. & Harris, A. L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 491, 364–373 (2012).
de Koning, T. J. et al. L-serine in disease and development. Biochem. J. 371, 653–661 (2003).
Kalhan, S. C. & Hanson, R. W. Resurgence of serine: an often neglected but indispensable amino acid. J. Biol. Chem. 287, 19786–19791 (2012).
Shane, B. & Stokstad, E. L. Vitamin B12-folate interrelationships. Annu. Rev. Nutr. 5, 115–141 (1985).
Stipanuk, M. H. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 24, 539–577 (2004).
Stover, P. J. Physiology of folate and vitamin B12 in health and disease. Nutr. Rev. 62, S3–S13 (2004).
Farber, S. et al. The action of pteroylglutamic conjugates on man. Science 106, 619–621 (1947).
Farber, S., Diamond, L. K., Mercer, R.D., Sylvester, R. F. & Wolff, J. A. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. New Engl. J. Med. 238, 787–793 (1948). This pioneering study by Sydney Farber and colleagues provided proof of concept of cancer chemotherapy. They demonstrated that chemically targeting folate metabolism could induce remissions in patients with leukaemia.
Chabner, B. A. & Roberts, T. G. Jr. Chemotherapy and the war on cancer. Nature Rev. Cancer 5, 65–72 (2005).
Johnston, P. G. et al. Antimetabolites. Cancer Chemother. Biol. Response Modif. 17, 1–39 (1997).
Chabner, B. A., Myers, C. E., Coleman, C. N. & Johns, D. G. The clinical pharmacology of antineoplastic agents (second of two parts). New Engl. J. Med. 292, 1159–1168 (1975).
Chabner, B. A., Myers, C. E., Coleman, C. N. & Johns, D. G. The clinical pharmacology of antineoplastic agents (first of two parts). New Engl. J. Med. 292, 1107–1113 (1975).
McKnight, S. L. On getting there from here. Science 330, 1338–1339 (2010).
DeBerardinis, R. J. & Cheng, T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324 (2010).
Locasale, J. W. The consequences of enhanced cell-autonomous glucose metabolism. Trends Endocrinol. Metab. 23, 545–551 (2012).
Koppenol, W. H., Bounds, P. L. & Dang, C. V. Otto Warburg's contributions to current concepts of cancer metabolism. Nature Rev. Cancer 11, 325–337 (2011).
DeBerardinis, R. J. Serine metabolism: some tumors take the road less traveled. Cell. Metab. 14, 285–286 (2011).
Birsoy, K., Sabatini, D. M. & Possemato, R. Untuning the tumor metabolic machine: targeting cancer metabolism: a bedside lesson. Nature Med. 18, 1022–1023 (2012).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Rev. Mol. Cell Biol. 12, 21–35 (2011).
Wallingford, J. B., Niswander, L. A., Shaw, G. M. & Finnell, R. H. The continuing challenge of understanding, preventing, and treating neural tube defects. Science 339, http://dx.doi.org/10.1126/science.1222002 (2013).
Iskandar, B. J. et al. Folate regulation of axonal regeneration in the rodent central nervous system through DNA methylation. J. Clin. Invest. 120, 1603–1616 (2010).
Roth, C. et al. Folic acid supplements in pregnancy and severe language delay in children. JAMA 306, 1566–1573 (2011).
Wang, J. et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435–439 (2009).
Vander Heiden, M. G. et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harbor Symp. Quant. Biol. 76, 325–334 (2011).
Deberardinis, R. J., Sayed, N., Ditsworth, D. & Thompson, C. B. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18, 54–61 (2008).
Finkelstein, J. D. Methionine metabolism in mammals. J. Nutrit. Biochem. 1, 228–237 (1990).
Tong, X., Zhao, F. & Thompson, C. B. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr. Opin. Genet. Dev. 19, 32–37 (2009).
Gumaa, K. A., MacLeod, R. M. & McLean, P. The pentose phosphate pathway of glucose metabolism. Influence of a growth-hormone-secreting pituitary tumour on the oxidative and non-oxidative reactions of the cycle in liver. Biochem. J. 113, 215–220 (1969).
Zatz, M., Dudley, P. A., Kloog, Y. & Markey, S. P. Nonpolar lipid methylation. Biosynthesis of fatty acid methyl esters by rat lung membranes using S-adenosylmethionine. J. Biol. Chem. 256, 10028–10032 (1981).
Spector, A. A. & Yorek, M. A. Membrane lipid composition and cellular function. J. Lipid Res. 26, 1015–1035 (1985).
Aveldano, M. I. & Bazan, N. G. Molecular species of phosphatidylcholine, -ethanolamine, -serine, and -inositol in microsomal and photoreceptor membranes of bovine retina. J. Lipid Res. 24, 620–627 (1983).
Kinney, A. J. & Moore, T. S. Phosphatidylcholine Synthesis in Castor Bean Endosperm: I. Metabolism of l-Serine. Plant Physiol. 84, 78–81 (1987).
Hickman, M. J. et al. Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast. Mol. Biol. Cell 22, 4192–4204 (2011).
Rowe, P. B. & Lewis, G. P. Mammalian folate metabolism. Regulation of folate interconversion enzymes. Biochemistry 12, 1862–1869 (1973).
Bennett, B. D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature Chem. Biol. 5, 593–599 (2009).
Bennett, B. D., Yuan, J., Kimball, E. H. & Rabinowitz, J. D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nature Protoc. 3, 1299–1311 (2008).
Paul, B. D. & Snyder, S. H. H2S signalling through protein sulfhydration and beyond. Nature Rev. Mol. Cell Biol. 13, 499–507 (2012).
Yun, J., Johnson, J. L., Hanigan, C. L. & Locasale, J. W. Interactions between epigenetics and metabolism in cancers. Front. Oncol. 2, 163 (2012).
Anderson, O. S., Sant, K. E. & Dolinoy, D. C. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J. Nutrit. Biochem. 23, 853–859 (2012).
Katada, S., Imhof, A. & Sassone-Corsi, P. Connecting threads: epigenetics and metabolism. Cell 148, 24–28 (2012).
Gluckman, P. D. Epigenetics and metabolism in 2011: epigenetics, the life-course and metabolic disease. Nature Rev. Endocrinol. 8, 74–76 (2012).
Teperino, R., Schoonjans, K. & Auwerx, J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell. Metab. 12, 321–327 (2010).
Chen, C., Nott, T. J., Jin, J. & Pawson, T. Deciphering arginine methylation: Tudor tells the tale. Nature Rev. Mol. Cell Biol. 12, 629–642 (2011).
Yang, Y. & Bedford, M. T. Protein arginine methyltransferases and cancer. Nature Rev. Cancer 13, 37–50 (2013).
Chiang, P. K. et al. S-Adenosylmethionine and methylation. FASEB J. 10, 471–480 (1996).
Heby, O. & Persson, L. Molecular genetics of polyamine synthesis in eukaryotic cells. Trends Biochem. Sci. 15, 153–158 (1990).
Snyder, S. H. & Russell, D. H. Polyamine synthesis in rapidly growing tissues. Fed. Proc. 29, 1575–1582 (1970).
Janne, J., Raina, A. & Siimes, M. Mechanism of stimulation of polyamine synthesis by growth hormone in rat liver. Biochim. Biophys. Acta 166, 419–426 (1968).
Farber, S. Some observations on the effect of folic acid antagonists on acute leukemia and other forms of incurable cancer. Blood 4, 160–167 (1949).
Vogelzang, N. J. et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. 21, 2636–2644 (2003).
Vander Heiden, M. G. Targeting cancer metabolism: a therapeutic window opens. Nature Rev. Drug Discov. 10, 671–684 (2011).
Cheson, B. D. New antimetabolites in the treatment of human malignancies. Seminars Oncol. 19, 695–706 (1992).
Bertino, J. R. & New York Academy of Sciences. Section of Biological and Medical Sciences. Folate Antagonists As Chemotherapeutic Agents. (New York Academy of Sciences, 1971).
Sirotnak, F. M. Folate Antagonists As Therapeutic Agents. (Academic Press, 1984).
Baram, J., Allegra, C. J., Fine, R. L. & Chabner, B. A. Effect of methotrexate on intracellular folate pools in purified myeloid precursor cells from normal human bone marrow. J. Clin. Invest. 79, 692–697 (1987).
Allegra, C. J., Fine, R. L., Drake, J. C. & Chabner, B. A. The effect of methotrexate on intracellular folate pools in human MCF-7 breast cancer cells. Evidence for direct inhibition of purine synthesis. J. Biol. Chem. 261, 6478–6485 (1986).
Diasio, R. B. & Harris, B. E. Clinical pharmacology of 5-fluorouracil. Clin. Pharmacokinet. 16, 215–237 (1989).
Pinedo, H. M. & Peters, G. F. Fluorouracil: biochemistry and pharmacology. J. Clin. Oncol. 6, 1653–1664 (1988).
Spears, C. P., Shahinian, A. H., Moran, R. G., Heidelberger, C. & Corbett, T. H. In vivo kinetics of thymidylate synthetase inhibition of 5-fluorouracil-sensitive and -resistant murine colon adenocarcinomas. Cancer Res. 42, 450–456 (1982).
Burris, H. A. et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 15, 2403–2413 (1997).
Plunkett, W. et al. Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Seminars Oncol. 22, 3–10 (1995).
Heinemann, V., Schulz, L., Issels, R. D. & Plunkett, W. Gemcitabine: a modulator of intracellular nucleotide and deoxynucleotide metabolism. Seminars Oncol. 22, 11–18 (1995).
Bouffard, D. Y., Laliberte, J. & Momparler, R. L. Kinetic studies on 2′,2′-difluorodeoxycytidine (Gemcitabine) with purified human deoxycytidine kinase and cytidine deaminase. Biochem. Pharmacol. 45, 1857–1861 (1993).
Jones, P. A. & Baylin, S. B. The epigenomics of cancer. Cell 128, 683–692 (2007).
Yoo, C. B. & Jones, P. A. Epigenetic therapy of cancer: past, present and future. Nature Rev. Drug Discov. 5, 37–50 (2006).
Egger, G., Liang, G., Aparicio, A. & Jones, P. A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 (2004).
Rodriguez-Paredes, M. & Esteller, M. Cancer epigenetics reaches mainstream oncology. Nature Med. 17, 330–339 (2011).
Chi, P., Allis, C. D. & Wang, G. G. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature Rev. Cancer 10, 457–469 (2010).
Stresemann, C., Brueckner, B., Musch, T., Stopper, H. & Lyko, F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 66, 2794–2800 (2006).
Lyko, F. & Brown, R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J. Natl Cancer Institute 97, 1498–1506 (2005).
Spannhoff, A., Hauser, A. T., Heinke, R., Sippl, W. & Jung, M. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem 4, 1568–1582 (2009).
Tsai, H. C. et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21, 430–446 (2012).
Wagner, T. & Jung, M. New lysine methyltransferase drug targets in cancer. Nature Biotechnol. 30, 622–623 (2012).
Goldberg, A. D., Allis, C. D. & Bernstein, E. Epigenetics: a landscape takes shape. Cell 128, 635–638 (2007).
Zhang, Y. & Reinberg, D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360 (2001).
Brueckner, B., Kuck, D. & Lyko, F. DNA methyltransferase inhibitors for cancer therapy. Cancer J. 13, 17–22 (2007).
Yao, Y. et al. Selective inhibitors of histone methyltransferase DOT1L: design, synthesis, and crystallographic studies. J. Am. Chem. Soc. 133, 16746–16749 (2011).
Casero, R. A. Jr & Marton, L. J. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nature Rev. Drug Discov. 6, 373–390 (2007).
Snell, K. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv. Enzyme Regul. 22, 325–400 (1984).
Fell, D. A. & Snell, K. Control analysis of mammalian serine biosynthesis. Feedback inhibition on the final step. Biochem. J. 256, 97–101 (1988).
Snell, K., Natsumeda, Y. & Weber, G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem. J. 245, 609–612 (1987).
Snell, K. & Weber, G. Enzymic imbalance in serine metabolism in rat hepatomas. Biochem. J. 233, 617–620 (1986).
Kit, S. The biosynthesis of free glycine and serine by tumors. Cancer Res. 15, 715–718 (1955).
Locasale, J. W. & Cantley, L. C. Genetic selection for enhanced serine metabolism in cancer development. Cell Cycle 10, 3812–3813 (2011).
Locasale, J. W. et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genet. 43, 869–874 (2011).
Possemato, R. et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 (2011). References 90 and 91 showed that de novo serine biosynthesis through PHGDH was dramatically enhanced in a subset of breast cancers and melanomas. Those with copy number gains selectively depended on PHGDH for proliferation, supporting the hypothesis that PHGDH is a candidate oncogene.
Pollari, S. et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res. Treat. 125, 421–430 (2011).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Chaneton, B. et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491, 458–462 (2012).
Hitosugi, T. et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell 22, 585–600 (2012).
Kung, C. et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem. Biol. 19, 1187–1198 (2012).
Ye, J. et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc. Natl Acad. Sci. USA 109, 6904–6909 (2012).
Maddocks, O. D. et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546 (2013). This study demonstrated the dietary requirements of serine and glycine for in vivo tumour growth, as well as the p53 dependency of cancer cells to adapt to serine and glycine deprivation.
Ma, L. et al. Control of nutrient stress-induced metabolic reprogramming by PKCzeta in tumorigenesis. Cell 152, 599–611 (2013).
Zhang, W. C. et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148, 259–272 (2012). This study demonstrated that increased GLDC activity was found in a subset of tumour-initiating cells and that increased GLDC activity was sufficient to induce tumour growth in a mouse xenograft.
Wang, J., Alexander, P. & McKnight, S. L. Metabolic specialization of mouse embryonic stem cells. Cold Spring Harbor Symp. Quant. Biol. 76, 183–193 (2011).
Alexander, P. B., Wang, J. & McKnight, S. L. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc. Natl Acad. Sci. USA 108, 15828–15833 (2011).
Shyh-Chang, N. et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226 (2013). This study demonstrated the ability of changes in the levels of metabolites in one-carbon metabolism to regulate the levels of histone methylation.
Ramm Sander, P. et al. Stem cell metabolic and spectroscopic profiling. Trends Biotechnol. 31, 204–213 (2013).
Sassone-Corsi, P. Physiology. When metabolism and epigenetics converge. Science 339, 148–150 (2013).
Jain, M. et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 (2012). This study carried out a metabolic flux analysis across the panel of NCI-60 cell lines. It was found that glycine uptake was the flux that most strongly correlated with cell proliferation.
Sreekumar, A. et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 (2009). This study carried out a metabolomics analysis of urine from a large cohort of patients with prostate cancer. The studies identified glycine and sarcosine as potential predictors of metastatic prostate cancer.
Scuoppo, C. et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature 487, 244–248 (2012). This study identified genes involved in polyamine metabolism as having a tumour-suppressive role in oncogenesis. Polyamines are derived from S -adenosylmethione.
Igarashi, K. & Kashiwagi, K. Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 42, 39–51 (2010).
Park, M. H., Nishimura, K., Zanelli, C. F. & Valentini, S. R. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids 38, 491–500 (2010).
Saini, P., Eyler, D. E., Green, R. & Dever, T. E. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121 (2009).
Lee, S. B. et al. The effect of hypusine modification on the intracellular localization of eIF5A. Biochem. Biophys. Res. Commun. 383, 497–502 (2009).
Park, M. H. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J. Biochem. 139, 161–169 (2006).
Silvera, D., Formenti, S. C. & Schneider, R. J. Translational control in cancer. Nature Rev. Cancer 10, 254–266 (2010).
Hoeijmakers, J. H. Genome maintenance mechanisms are critical for preventing cancer as well as other aging-associated diseases. Mech. Ageing Dev. 128, 460–462 (2007).
Hoeijmakers, J. H. Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 (2001).
Hu, C. M. et al. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer Cell 22, 36–50 (2012).
Wilson, P. M., LaBonte, M. J., Lenz, H. J., Mack, P. C. & Ladner, R. D. Inhibition of dUTPase induces synthetic lethality with thymidylate synthase-targeted therapies in non-small cell lung cancer. Mol. Cancer Ther. 11, 616–628 (2012).
Stover, P. J. & Weiss, R. S. Sensitizing cancer cells: is it really all about U? Cancer Cell 22, 3–4 (2012).
Bester, A. C. et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 (2011). This study showed that changes in the levels of nucleotide intermediates that are derived from one-carbon metabolism are sufficient to induce tumorigenesis.
Ohta, M. et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 84, 587–597 (1996). This study identified a tumour suppressor gene, FHIT , which encodes a metabolic enzyme involved in nucleotide metabolism, as being recurrently deleted in multiple cancers, including colorectal cancer.
Sozzi, G. et al. The FHIT gene 3p14.2 is abnormal in lung cancer. Cell 85, 17–26 (1996).
Pekarsky, Y., Zanesi, N., Palamarchuk, A., Huebner, K. & Croce, C. M. FHIT: from gene discovery to cancer treatment and prevention. Lancet Oncol. 3, 748–754 (2002).
Saldivar, J. C. et al. Initiation of genome instability and preneoplastic processes through loss of Fhit expression. PLoS Genet. 8, e1003077 (2012).
Siprashvili, Z. et al. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc. Natl Acad. Sci. USA 94, 13771–13776 (1997).
Rahman, L. et al. Thymidylate synthase as an oncogene: a novel role for an essential DNA synthesis enzyme. Cancer Cell 5, 341–351 (2004). This study provided evidence that thymidylate synthase can function as a metabolic oncogene.
Fukushima, M., Fujioka, A., Uchida, J., Nakagawa, F. & Takechi, T. Thymidylate synthase (TS) and ribonucleotide reductase (RNR) may be involved in acquired resistance to 5-fluorouracil (5-FU) in human cancer xenografts in vivo. Eur. J. Cancer 37, 1681–1687 (2001).
Takezawa, K. et al. Identification of thymidylate synthase as a potential therapeutic target for lung cancer. Br. J. Cancer 103, 354–361 (2010).
Xu, X. et al. Broad overexpression of ribonucleotide reductase genes in mice specifically induces lung neoplasms. Cancer Res. 68, 2652–2660 (2008).
Bertino, J. R. & Banerjee, D. Thymidylate synthase as an oncogene? Cancer Cell 5, 301–302 (2004).
Jones, P. A. & Takai, D. The role of DNA methylation in mammalian epigenetics. Science 293, 1068–1070 (2001).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nature Rev. Mol. Cell Biol. 6, 838–849 (2005).
Delhommeau, F. et al. Mutation in TET2 in myeloid cancers. New Engl. J. Med. 360, 2289–2301 (2009).
Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. New Engl. J. Med. 363, 2424–2433 (2010).
Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010).
Ceol, C. J. et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–517 (2011).
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Zee, B. M. et al. In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 285, 3341–3350 (2010).
Barth, T. K. & Imhof, A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 35, 618–626 (2010).
Wang, G. G., Allis, C. D. & Chi, P. Chromatin remodeling and cancer, Part I: covalent histone modifications. Trends Mol. Med. 13, 363–372 (2007).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Shaw, R. J. & Cantley, L. C. Decoding key nodes in the metabolism of cancer cells: sugar and spice and all things nice. F1000 Biol. Rep. 4, 2 (2012).
Cheong, H., Lu, C., Lindsten, T. & Thompson, C. B. Therapeutic targets in cancer cell metabolism and autophagy. Nature Biotechnol. 30, 671–678 (2012).
Teicher, B. A., Linehan, W. M. & Helman, L. J. Targeting cancer metabolism. Clin. Cancer Res. 18, 5537–5545 (2012).
Dann, S. G. & Abraham, R. T. Serine biosynthesis: fuel for the melanoma cell growth engine. Pigment Cell Melanoma Res. 24, 875–877 (2011).
Trachootham, D., Alexandre, J. & Huang, P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature Rev. Drug Discov. 8, 579–591 (2009).
Inzucchi, S. E. et al. Efficacy and metabolic effects of metformin and troglitazone in type II diabetes mellitus. New Engl. J. Med. 338, 867–872 (1998).
Cabreiro, F. et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 (2013).
Jalving, M. et al. Metformin: taking away the candy for cancer? Eur. J. Cancer 46, 2369–2380 (2010).
Pierotti, M. A. et al. Targeting metabolism for cancer treatment and prevention: metformin, an old drug with multi-faceted effects. Oncogene 32, 1475–1487 (2013).
Chong, C. R. & Chabner, B. A. Mysterious metformin. Oncol. 14, 1178–1181 (2009).
Corominas-Faja, B. et al. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging 4, 480–498 (2012).
Fedirko, V. et al. Glycemic index, glycemic load, dietary carbohydrate, and dietary fiber intake and risk of liver and biliary tract cancers in Western Europeans. Ann. Oncol. 24, 543–553 (2013).
Nagle, C. M. et al. Carbohydrate intake, glycemic load, glycemic index, and risk of ovarian cancer. Ann. Oncol. 22, 1332–1338 (2011).
Patel, A. V. et al. Glycemic load, glycemic index, and carbohydrate intake in relation to pancreatic cancer risk in a large US cohort. Cancer Causes Control 18, 287–294 (2007).
Oh, K., Willett, W. C., Fuchs, C. S. & Giovannucci, E. L. Glycemic index, glycemic load, and carbohydrate intake in relation to risk of distal colorectal adenoma in women. Cancer Epidemiol. Biomarkers Prev. 13, 1192–1198 (2004).
Ho, V. W. et al. A low carbohydrate, high protein diet slows tumor growth and prevents cancer initiation. Cancer Res. 71, 4484–4493 (2011).
Myers, A. P. & Cantley, L. C. Sugar free, cancer free? Nutrition 28, 1036 (2012).
Fine, E. J. et al. Targeting insulin inhibition as a metabolic therapy in advanced cancer: a pilot safety and feasibility dietary trial in 10 patients. Nutrition 28, 1028–1035 (2012).
Tavana, O. & Gu, W. The hunger games: p53 Regulates metabolism upon serine starvation. Cell. Metab. 17, 159–161 (2013).
Shackelford, D. B. & Shaw, R. J. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature Rev. Cancer 9, 563–575 (2009).
Wu, K. et al. A prospective study on folate, B12, and pyridoxal 5′-phosphate (B6) and breast cancer. Cancer Epidemiol. Biomarkers Prev. 8, 209–217 (1999).
Crider, K. S., Yang, T. P., Berry, R. J. & Bailey, L. B. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Adv. Nutr. 3, 21–38 (2012).
Guerreiro, C. S. et al. Risk of colorectal cancer associated with the C677T polymorphism in 5,10-methylenetetrahydrofolate reductase in Portuguese patients depends on the intake of methyl-donor nutrients. Am. J. Clin. Nutr. 88, 1413–1418 (2008).
Ma, J. et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 57, 1098–1102 (1997).
Giovannucci, E. et al. Folate, methionine, and alcohol intake and risk of colorectal adenoma. J. Natl Cancer Institute 85, 875–884 (1993).
van Engeland, M. et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 63, 3133–3137 (2003).
Figueiredo, J. C. et al. Folic acid and prevention of colorectal adenomas: a combined analysis of randomized clinical trials. Int. J. Cancer 129, 192–203 (2011).
Ibrahim, E. M. & Zekri, J. M. Folic acid supplementation for the prevention of recurrence of colorectal adenomas: metaanalysis of interventional trials. Med. Oncol. 27, 915–918 (2010).
Logan, R. F., Grainge, M. J., Shepherd, V. C., Armitage, N. C. & Muir, K. R. Aspirin and folic acid for the prevention of recurrent colorectal adenomas. Gastroenterology 134, 29–38 (2008).
Chae, Y. K. & Yun, J. H. Folic acid and prevention of colorectal adenomas. JAMA 298, 1397 (2007).
Cole, B. F. et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA 297, 2351–2359 (2007).
Christensen, B. C. et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 6, e1001043 (2010).
Zhang, S. et al. A prospective study of folate intake and the risk of breast cancer. JAMA 281, 1632–1637 (1999).
Homocysteine Lowering Trialists' Collaboration. Lowering blood homocysteine with folic acid based supplements: meta-analysis of randomised trials. BMJ 316, 894–898 (1998).
Shirota, Y. et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J. Clin. Oncol. 19, 4298–4304 (2001).
Metzger, R. et al. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J. Clin. Oncol. 16, 309–316 (1998).
Edler, D. et al. Thymidylate synthase expression in colorectal cancer: a prognostic and predictive marker of benefit from adjuvant fluorouracil-based chemotherapy. J. Clin. Oncol. 20, 1721–1728 (2002).
Popat, S., Matakidou, A. & Houlston, R. S. Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta-analysis. J. Clin. Oncol. 22, 529–536 (2004).
Walther, A. et al. Genetic prognostic and predictive markers in colorectal cancer. Nature Rev. Cancer 9, 489–499 (2009).
Dervieux, T., Greenstein, N. & Kremer, J. Pharmacogenomic and metabolic biomarkers in the folate pathway and their association with methotrexate effects during dosage escalation in rheumatoid arthritis. Arthritis Rheum. 54, 3095–3103 (2006).
Valik, D., Radina, M., Sterba, J. & Vojtesek, B. Homocysteine: exploring its potential as a pharmacodynamic biomarker of antifolate chemotherapy. Pharmacogenomics 5, 1151–1162 (2004).
Vazquez, A., Tedeschi, P. M. & Bertino, J. R. Overexpression of the mitochondrial folate and glycine-serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer Res. 73, 478–482 (2013).
Jentzmik, F. et al. Sarcosine in urine after digital rectal examination fails as a marker in prostate cancer detection and identification of aggressive tumours. Eur. Urol. 58, 12–18 (2010).
Ploussard, G. & de la Taille, A. Urine biomarkers in prostate cancer. Nature Rev. Urol. 7, 101–109 (2010).
Couzin, J. Biomarkers. Metabolite in urine may point to high-risk prostate cancer. Science 323, 865 (2009).
Glunde, K., Bhujwalla, Z. M. & Ronen, S. M. Choline metabolism in malignant transformation. Nature Rev. Cancer 11, 835–848 (2011).
Aboagye, E. O. & Bhujwalla, Z. M. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 59, 80–84 (1999).
Hara, T., Kosaka, N. & Kishi, H. PET imaging of prostate cancer using carbon-11-choline. J. Nucl. Med. 39, 990–995 (1998).
Reske, S. N. et al. Imaging prostate cancer with 11C-choline PET/CT. J. Nucl. Med. 47, 1249–1254 (2006).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. New Engl. J. Med. 351, 2817–2826 (2004).
Dang, L. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009).
Losman, J. A. & Kaelin, W. G. Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 (2013).
Kaelin, W. G. Jr & McKnight, S. L. Influence of metabolism on epigenetics and disease. Cell 153, 56–69 (2013).
Chu, E. Ode to 5-Fluorouracil. Clin. Colorectal Cancer 6, 609 (2007).
Acknowledgements
The author is grateful to members of his laboratory for helpful discussions. He also thanks L. Cantley for stimulating conversations in this area and the anonymous reviewers for their helpful comments. The work was supported through funding from the American Cancer Society and the US National Cancer Institute (CA168997).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Glossary
- Aldol cleavage
-
A chemical reaction that can be catalysed enzymatically resulting in the splitting of a β-hydroxy ketone.
- NCI-60
-
A panel of tumour-derived cell lines originating from diverse tissue types. Extensive genomic, biochemical and pharmacological data have been obtained on these cell lines.
- Therapeutic window
-
A term used in drug development and medical practice that refers to ranges of drug concentrations that satisfy the trade-off between an efficacious clinical response and unwanted toxicities.
Rights and permissions
About this article
Cite this article
Locasale, J. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13, 572–583 (2013). https://doi.org/10.1038/nrc3557
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3557
This article is cited by
-
Comprehensive analysis the prognostic and immune characteristics of mitochondrial transport-related gene SFXN1 in lung adenocarcinoma
BMC Cancer (2024)
-
Immune, metabolic landscapes of prognostic signatures for lung adenocarcinoma based on a novel deep learning framework
Scientific Reports (2024)
-
Targeting serine/glycine metabolism improves radiotherapy response in non-small cell lung cancer
British Journal of Cancer (2024)
-
Current and Potential Roles of Ferroptosis in Bladder Cancer
Current Medical Science (2024)
-
Identification of glucose-independent and reversible metabolic pathways associated with anti-proliferative effect of metformin in liver cancer cells
Metabolomics (2024)